
ΑΙhub.org
#ICML2022 invited talk round-up 2: estimating causal effects and drug discovery and development

In this post, we summarise the final two invited talks from the International Conference on Machine Learning (ICML 2022). These presentations covered estimation and inference for causal effects, and machine learning for drug discovery and development.
Synthetic control methods and difference-in-differences – Guido Imbens
Guido’s talk covered the topic of estimation and inference for causal effects in panel data settings, in particular focussing on synthetic control methods and difference-in-difference methods. These methods are very popular in the empirical literature in economics, but many questions remain concerning causal effects in these settings. There has been a lot of recent theoretical work trying to improve practices in this field.
Guido devoted most of the presentation to explaining how one might go about estimating the causal effect of some treatment, with a specific focus on binary treatments. The particular setting that he looked at is one where we observe multiple units (these units could be individuals, states, countries, or companies, for example) repeatedly over time as a particular treatment is administered. In some problems, the number of units is large and they are observed over a short time period. In others, the number of units is small, but they are observed over a long time period.
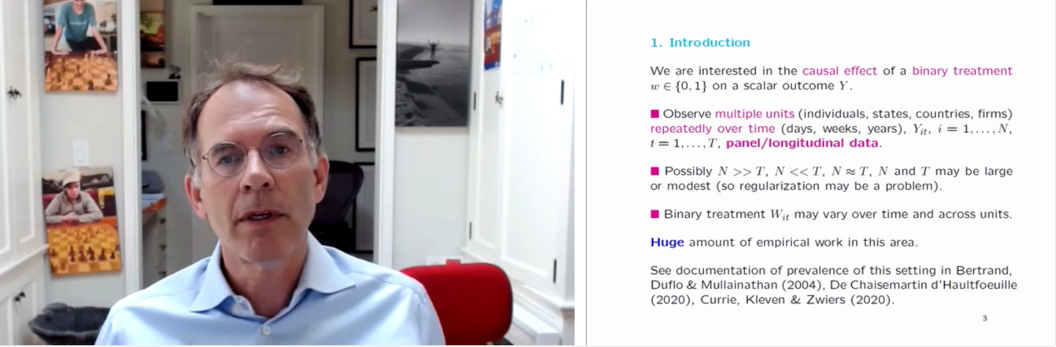 Screenshot from Guido’s talk.
Screenshot from Guido’s talk.
Firstly, Guido focussed on the case where there are two observations for each unit and time period: one observation is the outcome, and the other observation is the binary treatment. The units are split into a treatment group and a control group, and we are interested in the average difference between the treated group and the control group. Guido gave some examples of where these methods have been used in the past to evaluate the effect of treatments. These include California smoking regulation, the re-unification of Germany, and minimum wage changes.
Lots of different methods have been used for estimating average treatment effects, including matching, inverse propensity score weighting, and double robust methods. Guido took the audience through some of the regression techniques used, looking at some different scenarios. He talked about the importance of selecting the method that is appropriate the application in question, and looking for combinations of different methods to find the most effective.
To close, Guido left the audience with one question for future research: how can we systematically combine experimental and observational data to answer questions that neither can directly answer?
To find out more, you can see the slides from Guido’s talk here.
Design for inference in drug discovery and development – Aviv Regev
To introduce her talk, Aviv took us on a whistle-stop tour of the last 40 years of medical history, in which many technological developments, combined with biological insights, have enabled medical breakthroughs. These include recombinant DNA technologies which led to protein drugs, and therapeutic antibodies which have brought many important medicines.
Aviv’s research mission is to use machine learning to try to make fundamental scientific discoveries and to use these insights to help in the development of therapeutics for patients. One promising area for research is human genetics. Over the course of the last 15 years, analysis of the variation in the genomes of hundreds of thousands of individuals has identified about 100,000 regions in the human genome where differences in the DNA sequence are associated with the risk of developing a range of different diseases. Looking at this from the point-of-view of drug discovery, this analysis gives researchers a starting point as to where to begin in the search for potential new drugs.
 Screenshot from Aviv’s talk.
Screenshot from Aviv’s talk.
In her talk, Aviv shared some examples of how machine learning and experiments come together in target discovery, drug discovery, and clinical development. She primarily focussed on the first of these: target discovery.
Most of the genetic variants in our genome associated with common complex diseases are within regions that regulate gene expression. The question is: how can we progress from an understanding of the initial changes in the regulatory region to an understanding of the impact of those changes on genes and cells (i.e. the effect on our bodies)? Aviv outlined the method that she and her team use to try to answer this question. They start with a biological system (for example, the tissue or cell that plays an important role in the disease in question), they perturb it experimentally, then they measure the effect using high-resolution techniques. From these data, they train a machine learning model and query it for a hypothesis. They then use that hypothesis to design the next experiment. The process is repeated to design further experiments.
Aviv and her team have developed a deep transformer neural network model that takes any arbitrary DNA sequence and predicts the level of gene expression. The goal is that, with such a model, they can predict what a new variant does. This the first step in moving from human genetics towards a new drug. Training the model required a huge number of examples of regulatory sequences and the associated measured expression – more than currently exist. To get round this problem, the team used randomly generated DNA sequences for extra training examples. Because the model generalises extremely well, they could use it as an oracle to design sequences for gene expression engineering.
In the final part of her talk, Aviv touched on the next part of the process: drug discovery. She and her team have investigated combining machine learning models and experiments for studying antibodies, small molecules and RNA vaccines.
You can find out more on her website.
tags: ICML, ICML2022










