
ΑΙhub.org
Botanical time machines: AI is unlocking a treasure trove of data held in herbarium collections
 A herbarium specimen of Cheiranthera linearis (commonly known as finger-flower), collected in 1912 by Edwin James Semmens, former principal of the Victorian School of Forestry. Image credit: University of Melbourne.
A herbarium specimen of Cheiranthera linearis (commonly known as finger-flower), collected in 1912 by Edwin James Semmens, former principal of the Victorian School of Forestry. Image credit: University of Melbourne.
By Robert Turnbull, The University of Melbourne and Joanne Birch, The University of Melbourne
In 1770, after Captain Cook’s Endeavour struck the Great Barrier Reef and was held up for repairs, botanists Joseph Banks and Daniel Solander collected hundreds of plants.
One of those pressed plants is among 170,000 specimens in the herbarium at the University of Melbourne.
Worldwide, more than 395 million specimens are housed in herbaria. Together they comprise an unparalleled record of Earth’s plant and fungal life over time.
We wanted to find a better, faster way to tap into this wealth of information. Our new research describes the development and testing of a new AI-driven tool Hespi (short for “herbarium specimen sheet pipeline”). It has the potential to revolutionise access to biodiversity data and open up new avenues for research.
 The specimen sheet for spreading nut-heads (Epaltes australis), collected by Joseph Banks and Daniel Solander in 1770. (Note, the collection date was historically incorrectly written as 1776 on the specimen label). University of Melbourne Herbarium Collection
The specimen sheet for spreading nut-heads (Epaltes australis), collected by Joseph Banks and Daniel Solander in 1770. (Note, the collection date was historically incorrectly written as 1776 on the specimen label). University of Melbourne Herbarium CollectionThe digitisation challenge
To unlock the full potential of herbaria, institutions worldwide are striving to digitise them. This means photographing each specimen at high resolution and converting the information on its label into searchable digital data.
Once digitised, specimen records can be made available to the public through online databases such as the University of Melbourne Herbarium Collection Online. They are also fed into large biodiversity portals such as the Australasian Virtual Herbarium, the Atlas of Living Australia, or the Global Biodiversity Information Facility. These platforms make centuries of botanical knowledge accessible to researchers everywhere.
But digitisation is a monumental task. Large herbaria, such as the National Herbarium of New South Wales and the Australian National Herbarium have used high-capacity conveyor belt systems to rapidly image millions of specimens. Even with this level of automation, digitising the 1.15 million specimens at the National Herbarium of NSW took more than three years.
For smaller institutions without industrial-scale setups, the process is far slower. Staff, volunteers and citizen scientists photograph specimens and painstakingly transcribe their labels by hand.
At the current pace, many collections won’t be fully digitised for decades. This delay keeps vast amounts of biodiversity data locked away. Researchers in ecology, evolution, climate science and conservation urgently need access to large-scale, accurate biodiversity datasets. A faster approach is essential.
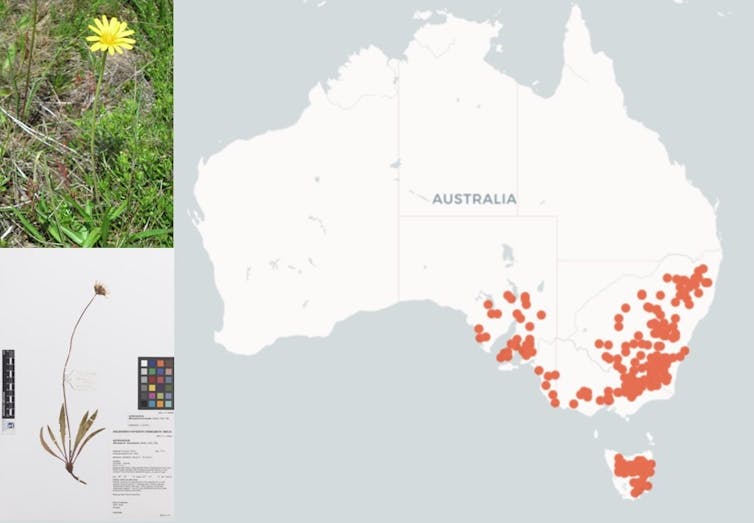 Map of specimen collection locations for Yam daisy (Microseris lanceolata) from records in The Australasian Virtual Herbarium.Neville Walsh, VicFlora
Map of specimen collection locations for Yam daisy (Microseris lanceolata) from records in The Australasian Virtual Herbarium.Neville Walsh, VicFloraHow AI is speeding things up
To address this challenge, we created Hespi – open-source software for automatically extracting information from herbarium specimens.
Hespi combines advanced computer vision techniques with AI tools such as object detection, image classification and large language models.
First, it takes an image of the specimen sheet which comprises the pressed plant and identifying text. Then it recognises and extracts text, using a combination of optical character recognition and handwritten text recognition.
Deciphering handwriting is challenging for people and computers alike. So Hespi passes the extracted text through OpenAI’s GPT-4o Large Language Model to correct any errors. This substantially improves the results.
So in seconds, Hespi locates the main specimen label on a herbarium sheet and reads the information it contains. This includes taxonomic names, collector details, location, latitude and longitude, and collection dates. It captures the data and converts it into a digital format, ready for use in research.
For example, Hespi correctly detected and extracted all relevant components from the herbarium sheet below. This large brown algae specimen was collected in 1883 at St Kilda.
 Results from Hespi on a sample of large brown algae (MELUA002557a) from the University of Melbourne, identifying important details such as the genus, species, locality and year of collection. University of Melbourne Herbarium
Results from Hespi on a sample of large brown algae (MELUA002557a) from the University of Melbourne, identifying important details such as the genus, species, locality and year of collection. University of Melbourne Herbarium
We tested Hespi on thousands of specimen images from the University of Melbourne Herbarium and other collections worldwide. We created test datasets for different stages in the pipeline and assessed the various components.
It achieved a high degree of accuracy. So it has the potential to save a lot of time, compared to manual data extraction.
We are developing a graphical user interface for the software so herbarium curators will be able to manually check and correct the results.
Just the beginning
Herbaria already contribute to society in many ways: from species identification and taxonomy to ecological monitoring, conservation, education, and even forensic investigations.
By mobilising large volumes of specimen-associated data, AI systems such as Hespi are enabling new and innovative applications at a scale never before possible.
AI has been used to automatically extract detailed leaf measurements and other traits from digitised specimens, unlocking centuries of historical collections for rapid research into plant evolution and ecology.
And this is just the beginning — computer vision and AI could soon be applied in many other ways, further accelerating and expanding botanical research in the years ahead.
 The digitisation pipeline at the University of Melbourne Herbarium begins with the generation of a high-resolution specimen image.Image credit: University of Melbourne Herbarium.
The digitisation pipeline at the University of Melbourne Herbarium begins with the generation of a high-resolution specimen image.Image credit: University of Melbourne Herbarium. Beyond herbaria
AI pipelines such as Hespi have the potential to extract text from labels in any museum or archival collection where high-quality digital images exist.
Our next step is a collaboration with Museums Victoria to adapt Hespi to create an AI digitisation pipeline suitable for museum collections. The AI pipeline will mobilise biodiversity data for about 12,500 specimens in the museum’s globally-significant fossil graptolite collection.
 A fossil graptolite specimen from Museums Victoria annotated by Hespi during data digitsition. Image credit: Museums Victoria.
A fossil graptolite specimen from Museums Victoria annotated by Hespi during data digitsition. Image credit: Museums Victoria.
We are also starting a new project with the Australian Research Data Commons (ARDC) to make the software more flexible. This will allow curators in museums and other institutions to customise Hespi to extract data from all kinds of collections — not just plant specimens.
Tranformational technology
Just as AI is reshaping many aspects of daily life, these technologies can transform access to biodiversity data. Human-AI collaborations could help overcome one of the biggest bottlenecks in collection digitisation — the slow, manual transcription of label data.
Mobilising the information already locked in herbaria, museums, and archives worldwide is essential to make it available for the cross-disciplinary research needed to understand and address the biodiversity crisis.
We wish to acknowledge our colleagues at the Melbourne Data Analytics Platform, including Karen Thompson and Emily Fitzgerald, who contributed to this research.![]()
Robert Turnbull, Senior Research Data Specialist, The University of Melbourne and Joanne Birch, Senior Lecturer in the School of BioSciences, and Herbarium Curator, The University of Melbourne
This article is republished from The Conversation under a Creative Commons license. Read the original article.










