
ΑΙhub.org
Discrete flow matching framework for graph generation
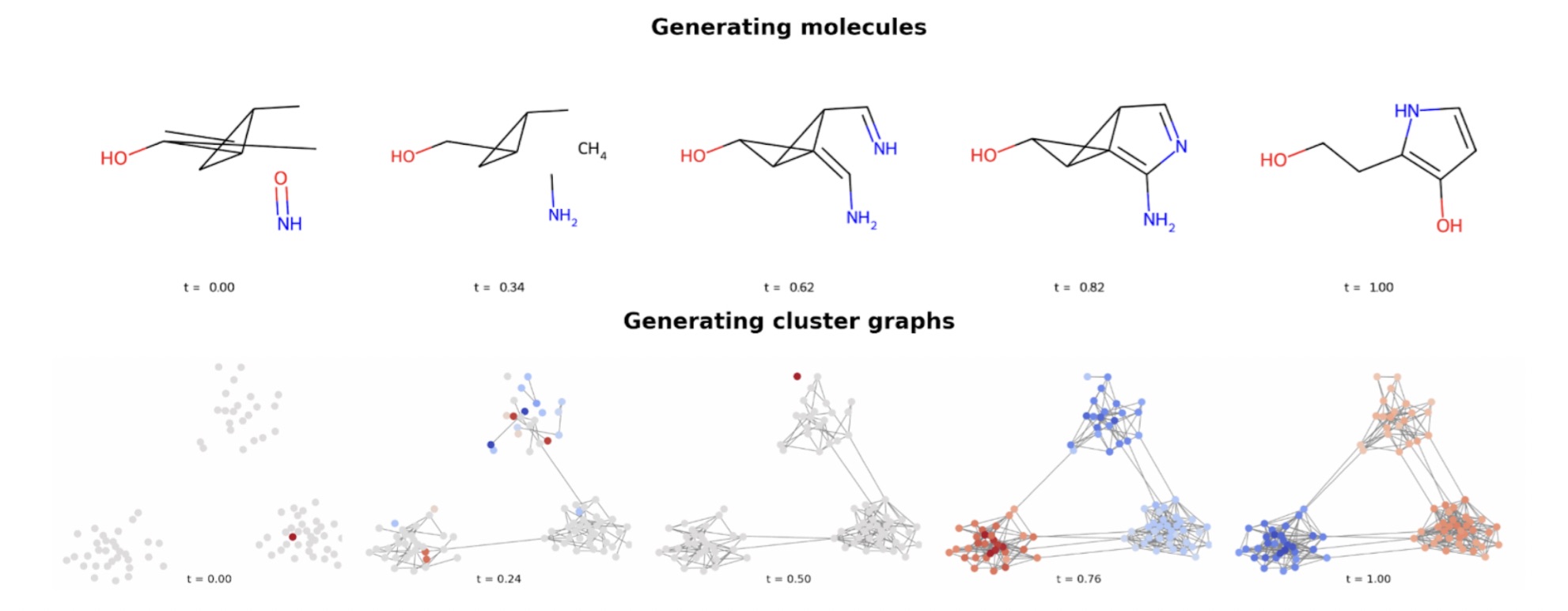 Figure 1: DeFoG progressively denoises graphs, transforming random structures (at t=0) into realistic ones (at t=1). The process is similar to reassembling scattered puzzle pieces back into their correct places.
Figure 1: DeFoG progressively denoises graphs, transforming random structures (at t=0) into realistic ones (at t=1). The process is similar to reassembling scattered puzzle pieces back into their correct places.
Designing a new drug often means inventing molecules that have never existed before. Chemists represent molecules as graphs, where atoms are the “nodes” and chemical bonds the “edges,” capturing their connections. This graph representation expands far beyond chemistry: a social network is a graph of people and friendships, the brain is a graph of neurons and synapses, and a transport system is a graph of stations and routes. From molecules to social networks, graphs are everywhere and naturally capture the relational structure of the world around us.
Therefore, for many applications, being able to generate new realistic graphs is a central problem. However, the scale of the problem is daunting: for example, a graph with 500 nodes could contain over 100,000 possible edges. Exploring such a vast combinatorial space by hand is impossible. This is why developing AI models capable of efficiently navigating this space and proposing thousands or even millions of new molecules, circuits, or networks in minutes would be a major scientific step forward.
Yet AI-based graph generation is far from straightforward. A particularly powerful family of approaches borrows ideas from image generation, especially diffusion models [1-3]. These models gradually add noise to a graph and then learn to reverse the process, a bit like shaking apart a completed jigsaw puzzle and reassembling it piece by piece (Figure 1). The main drawback is rigidity: the way a diffusion model is trained fixes the way it generates. This makes sampling slow, and if researchers want to generate more graphs, say 10,000 molecules instead of 1,000, this limitation quickly becomes a bottleneck. Even more challenging, adjusting the generation process to make it faster, slower, or tuned to a specific goal often requires retraining the entire model from scratch, which is one of the most computationally costly steps in the pipeline.
A new approach: DeFoG
At this year’s ICML conference, we introduced DeFoG, a discrete flow matching framework for graph generation [4]. Like diffusion models, DeFoG also progressively constructs a clean graph from a noisy one, but it does so in a more flexible formulation, based on discrete flow matching, which decouples training from generation. During training, the model focuses on a single skill: how to denoise, i.e., how to reverse a noisy graph back into a clean one. At generation time, however, DeFoG allows practitioners the freedom to decide how denoising unfolds. They can proceed more aggressively at the beginning and more cautiously at the end, or adapt the schedule in other ways to match the characteristics of the graphs at hand (see Figure 2). Much like choosing different routes on a map depending on whether you want the fastest, the safest, or the most scenic journey, this flexibility allows the generation process to better accommodate the characteristics of different families of graphs, such as the molecular and cluster graphs illustrated in Figure 1, leading to improved generative performance.
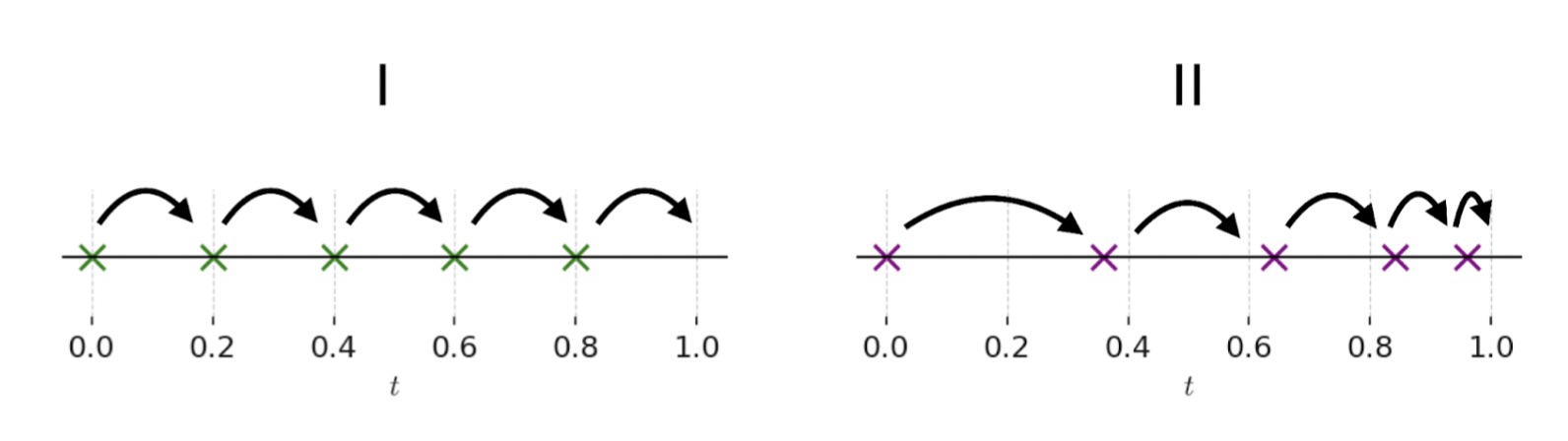 Figure 2: One example of the flexibility enabled by DeFoG. In I, the denoising schedule uses evenly spaced steps. In II, the schedule adapts the step sizes, taking larger steps early and smaller ones near the end, which allows for more refined generation at that stage. This freedom of DeFoG to design different denoising trajectories, alongside other ways of tailoring the process, leads to improved generative performance.
Figure 2: One example of the flexibility enabled by DeFoG. In I, the denoising schedule uses evenly spaced steps. In II, the schedule adapts the step sizes, taking larger steps early and smaller ones near the end, which allows for more refined generation at that stage. This freedom of DeFoG to design different denoising trajectories, alongside other ways of tailoring the process, leads to improved generative performance.
Why does it matter?
The improvement of DeFoG is two-fold. First, in terms of accuracy, DeFoG generates graphs that more closely resemble real ones than those produced by competing models. On synthetic benchmarks such as trees and community networks, it reached performance close to the best achievable. On molecular design benchmarks, it showed an outstanding ability to produce molecules that were novel, non-repeated, and chemically valid, meaning they satisfied established chemical rules. Second, in terms of efficiency, DeFoG achieved competitive results with existing graph generative models while requiring only 5 to 10% of the steps compared to many diffusion models [5,6].
Both aspects are crucial for real-world applications. In drug discovery, researchers must sift through millions of potential molecules, so realistic candidates save wasted effort while efficient sampling accelerates the entire search. In reinforcement learning, rapid generation of valid graphs is essential to provide quick feedback, allowing agents to learn faster. Therefore, the gains DeFoG provides in realism and efficiency are not just technical: they can make a practical difference.
Looking ahead
DeFoG represents not only a technical advance but also a conceptual step forward: it disentangles training from generation, opening new possibilities for iterative refinement in graph generation. Future directions include adaptive strategies that automatically adjust the denoising trajectory, as well as extensions to more complex and larger structures such as protein interaction networks or urban transport systems. At the same time, limitations remain in scaling to very large graphs and in balancing efficiency with fidelity, which highlight open challenges. Overall, the separation of training and generation paves the way for more efficient and effective graph generation, bringing the field closer to impactful real-world applications.
References
[1] Niu, Chenhao, et al. Permutation invariant graph generation via score-based generative modeling. International Conference on Artificial Intelligence and Statistics (2020)
[2] Jo, Jaehyeong, Seul Lee, and Sung Ju Hwang. Score-based generative modeling of graphs via the system of stochastic differential equations. International Conference on Machine Learning (2022)
[3] Vignac, Clement, et al. Digress: Discrete denoising diffusion for graph generation. International Conference on Learning Representations (2023)
[4] Qin, Yiming, et al. Defog: Discrete flow matching for graph generation. International Conference on Machine Learning (2025)
[5] Xu, Zhe, et al. Discrete-state continuous-time diffusion for graph generation. Advances in Neural Information Processing Systems (2024)
[6] Siraudin, Antoine, et al. Cometh: A continuous-time discrete-state graph diffusion model. ArXiv (2024).
tags: ICML, ICML2025











