
ΑΙhub.org
High-performance computing and AI team up for COVID-19 diagnostic imaging
The Confederation of Laboratories for Artificial Intelligence Research in Europe (CLAIRE) taskforce on AI & COVID-19 supported the creation of a research group focused on AI-assisted diagnosis of COVID-19 pneumonia. The first results demonstrate the great potential of AI-assisted diagnostic imaging. Furthermore, the impact of the taskforce work is much larger, and it embraces the cross-fertilisation of artificial intelligence (AI) and high-performance computing (HPC): a partnership with rocketing potential for many scientific domains.

Through several initiatives aimed at improving the knowledge of COVID-19, containing its diffusion, and limiting its effects, CLAIRE’s COVID-19 taskforce was able to organise 150 volunteer scientists, divided into seven groups covering different aspects of how AI could be used to tackle the pandemic. Emanuela Girardi, the co-coordinator of the CLAIRE taskforce on AI & COVID-19, supported the setup of a novel European group to study the diagnosis of COVID-19 pneumonia assisted by artificial intelligence. Professor Marco Aldinucci from the University of Torino led the group.
The AI and COVID-19 research group
The group is composed of fifteen researchers from six institutions, comprising expertise in complementary disciplines (radiomics, artificial intelligence, and high-performance computing).
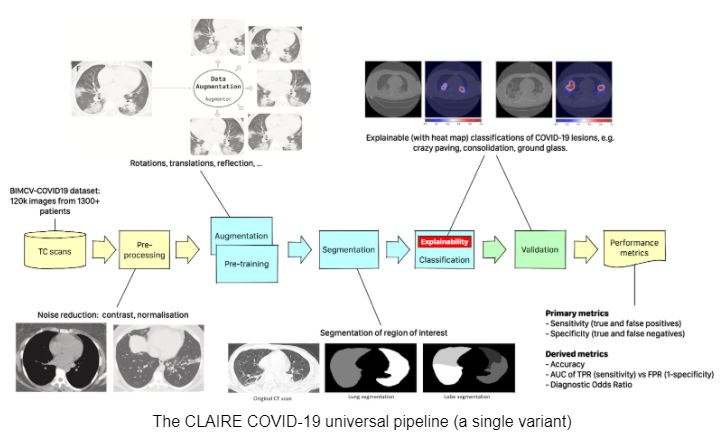
At the start of the pandemic, several research groups around the globe began to develop deep-learning models for the diagnosis of COVID-19. The use of different architectures, pipelines, and datasets makes it difficult to compare these models and identify the most promising ones. For this reason, the group set the task of defining a reproducible workflow capable of automating the comparison of state-of-the-art deep learning models for the diagnosis of COVID-19.
The workflow has subsequently evolved towards a “CLAIRE COVID-19 universal pipeline” for medical images. This pipeline, illustrated in the figure below, is capable of reproducing different state-of-the-art AI models already existing in the literature for the analysis of medical images. This provides a performance baseline for these techniques allowing the community to quantitatively measure the progress of AI in the diagnosis of COVID-19 and similar diseases. The “CLAIRE COVID-19 universal pipeline” includes innovative aspects, such as an “adversarial learning” step, that improves the interpretability of existing networks, and provides doctors not only with the diagnosis but also with indications of which lung lesions are crucial for the diagnosis.
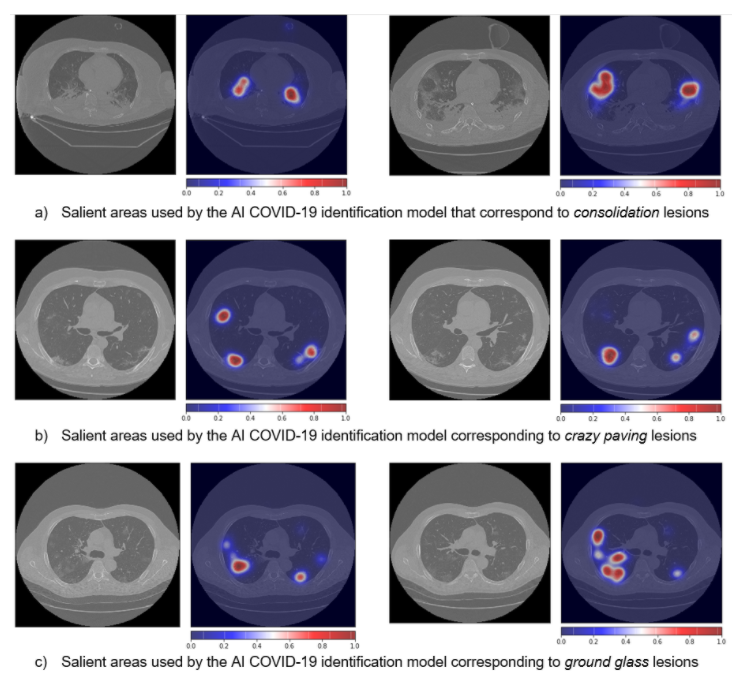
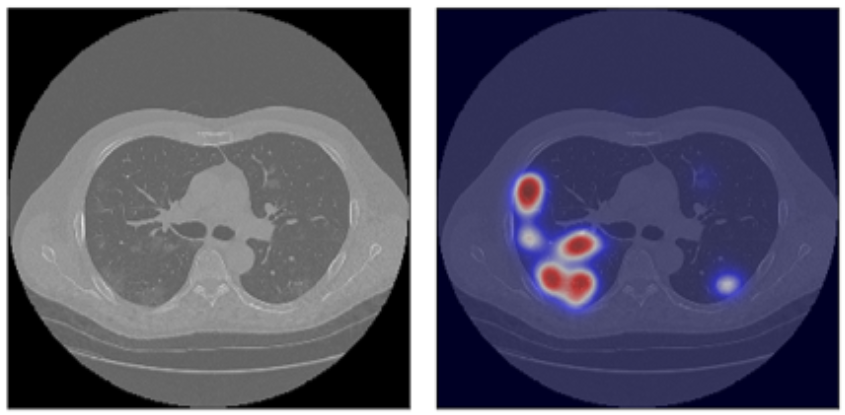
Interpretability of models is at the frontier of research in machine learning based on artificial neural networks (ANNs), which, although effective, remain obscure in providing explanations for their decisions. Typically, ANNs behave like black-boxes and it is difficult to explain why they have given a particular output. After processing a CT scan, an ANN will typically provide a decision (e.g., COVID-19 yes/no) with no explanation concerning the basis on which the decision was made.
The intensive use of neural networks means that the presence of high-performance computing experts in the group is beneficial. ANNs, used in their most effective form (called supervised learning), learn by digesting and generalising many human-labelled examples. In this case, many CT scans are labelled with a diagnosis made by an expert radiologist. The evolution of ANNs toward deep neural networks (DNNs) with multiple layers between the input and output layers, together with the capability to train them with an enormous number of examples has made it possible to improve the accuracy of the approach significantly. Both the accuracy and the usability of this approach are linked to the available computing power, which is enabled by hardware and software methods and tools in the area of high-performance computing (HPC).
The HPC-AI partnership
Theoretically, the CLAIRE COVID-19 universal pipeline is capable of reproducing the training of all the best models to diagnose pneumonia and to compare their performance. However, the adverb “theoretically” is strictly necessary. Moving from theory to practice requires two non-trivial ingredients to be found: a supercomputer of adequate computational power equipped with many latest generation GPUs and a mechanism capable of unifying and automating the execution of all variants of the workflow on a supercomputer. Training a single variant of the pipeline takes over 15 hours on a single high-end processor (e.g., NVidia V100 GPU); exploring all pipeline variants would take over two years. However, using a supercomputer could reduce this time down to one day.
Despite this potential, supercomputers are rarely used for AI. They are not yet equipped to effectively support specific AI software tools nor to acquire large amounts of data securely as required from medical field applications. Additionally, AI researchers are not used to the batch execution model used in supercomputers. Notwithstanding, the two communities need each other and are fated to meet. On the one hand, AI researchers need an ever-increasing data processing capability; on the other hand, supercomputers are shifting to GPUs because of their better energy efficiency and need for more and more GPU-enabled workloads, such as deep learning.
Scaling up development and benchmarking of AI systems will require significant amounts of computing power. Synergies between the AI and HPC communities should be leveraged to unlock these roadblocks. The work initiated by this group has led CLAIRE, ABD (Big Data Association), and CINI (National Interuniversity Consortium for Informatics) to sign a Memorandums of Understandings (MoU) aimed at providing substantial computing resources to the CLAIRE taskforce, and more generally, through CINI, to all Italian researchers active in research on COVID-19. Specifically, ABD, through its associate CINECA, has made available CINECA’s Marconi100 supercomputer. Marconi100 is currently ranked as 11th among the most powerful computers in the world. It is equipped with over 4000 NVidia V100 GPUs, and it is therefore particularly suitable for processing calculations for training deep neural networks (DNNs). Starting next year, thanks to the funding of 230 million euros obtained from the European program EuroHPC JU, CINECA will operate the new supercomputer “Leonardo” expected to be over ten times faster than Marconi100. Leonardo has been designed and is intended to be the most powerful machine in the world for AI applications (10 exaFLOPS FP16). Leonardo will also reach another record; it will be the first pre-exascale supercomputer designed and built in Europe (by Atos-BULL on CINECA specifications).
A second challenge for the taskforce has been the bridging of AI and HPC execution models. A problem approached describing the CLAIRE COVID-19 universal pipeline with Streamflow, an innovative Workflow Management System based on an open standard (CWL), which supports hybrid cloud-HPC deployments (via Kubernetes, SLURM, and other workload management systems). Streamflow, developed under the EU Deephealth and HPC4AI projects, directly addresses the two major obstacles to the use of supercomputers for AI applications:
- Usability: allows AI scientists to work with the standard tools of the research area provided via the cloud platform (for example, via Kubernetes) by automatically moving a large part of the calculation to the supercomputer used as a computing accelerator.
- Critical data: Streamflow moves data between cloud and HPC platforms, ensuring users have full control over who, how, and when to access data, without any waivers of trust to infrastructure managers.
The experiments have begun, and we are analysing the first encouraging results. The analysis of a small subset of the variants shows that the CLAIRE COVID-19 universal pipeline demonstrates the capability to generate models with excellent accuracy (over 90%) in classifying typical lesions of interstitial pneumonia due to COVID-19.
Despite the fact that the real impact of AI-assisted diagnosis on clinical practice remains to be assessed, the group, motivated by the COVID-19 emergency, has started to tackle a job that has the potential to pioneer the cross-fertilization of AI and HPC in the development of next-generation “Digital Twins”, so that no other pandemic can catch us unprepared.
About the group
The core members of the group are:
- Marco Aldinucci (coordinator), Iacopo Colonelli, Barbara Cantalupo – Parallel Computing Lab, University of Torino, Italy
- Marco Calandri, Piero Fariselli, Radiomics & medical science – University of Torino, Italy
- Marco Grangetto, Enzo Tartaglione – Digital image processing Lab, University of Torino, Italy
- Concetto Spampinato, Simone Palazzo, Isaak Kavasidis, Matteo Pennisi – PeRCeiVe Lab, University of Catania, Italy
- Bogdan Ionescu, Gabriel Constantin – Multimedia Lab @ CAMPUS Research Institute, University “Politehnica” of Bucharest, Romania
- Miquel Perello Nieto, Computer Science, University of Bristol, UK
- Inês Domingues, School of Sciences University of Porto, Portugal
tags: Focus on good health and well-being, Focus on UN SDGs










