
ΑΙhub.org
Putting nanoscale interactions under the microscope

By Lois Yoksoulian
Liquid-phase transmission electron microscopy (TEM) has recently been applied to materials chemistry to gain fundamental understanding of various reaction and phase transition dynamics at nanometer resolution. Researchers from the University of Illinois have developed a machine learning workflow to streamline the process of extracting physical and chemical parameters from TEM video data.
The new study, led by Qian Chen, a professor of materials science and engineering at the University of Illinois, Urbana-Champaign, builds upon her past work with liquid-phase electron microscopy and has been published in the journal ACS Central Science.
Being able to see – and record – the motions of nanoparticles is essential for understanding a variety of engineering challenges. Liquid-phase electron microscopy, which allows researchers to watch nanoparticles interact, is useful for research in medicine, energy and environmental sustainability and in fabrication of metamaterials, to name a few. However, it is difficult to interpret the dataset. The video files produced are large, filled with temporal and spatial information, and are noisy due to background signals – in other words, they require a lot of tedious image processing and analysis.
“Developing a method even to see these particles was a huge challenge,” Chen said. “Figuring out how to efficiently get the useful data pieces from a sea of outliers and noise has become the new challenge.”
To confront this problem, the team developed a machine learning workflow based on U-Net, a convolutional neural network that was developed for biomedical image segmentation. The workflow enables the researchers to achieve nanoparticle segmentation in liquid-phase TEM videos. U-Net can efficiently and precisely identify the boundary of nanoparticles.
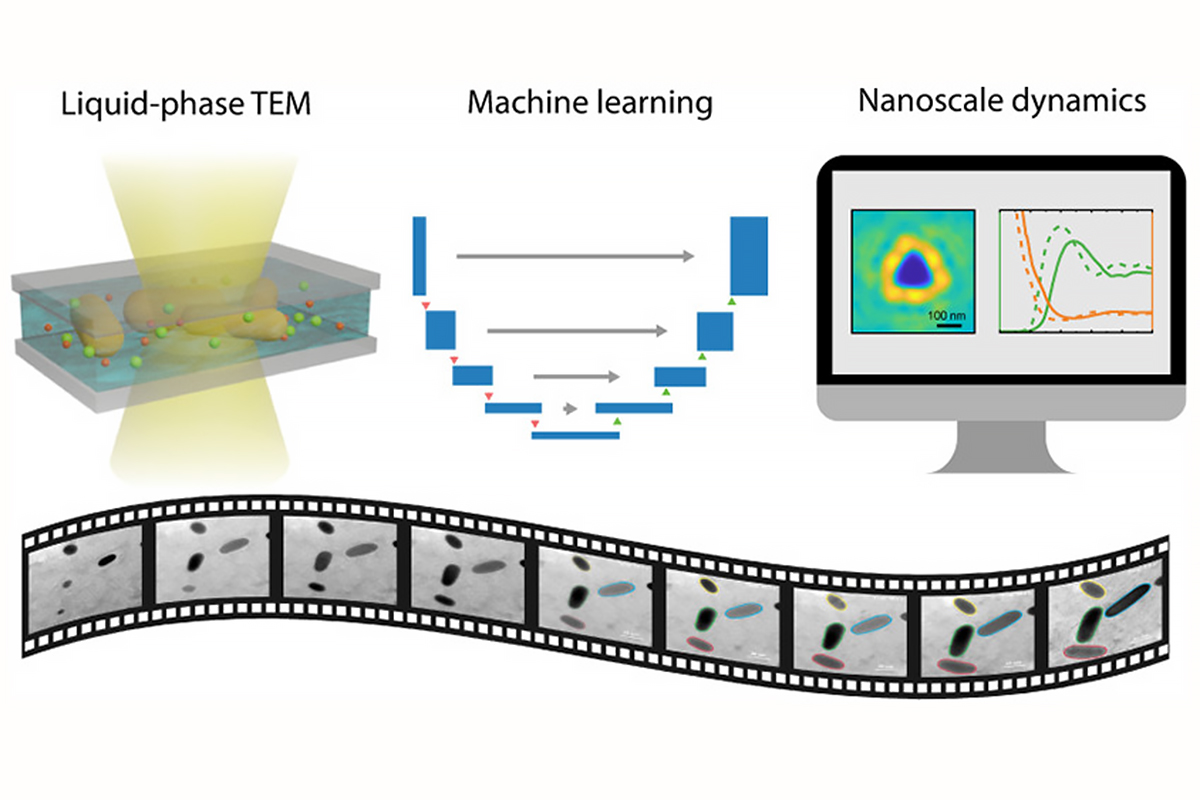
“Our new program processed information for three types of nanoscale dynamics including motion, chemical reaction and self-assembly of nanoparticles,” said lead author and graduate student Lehan Yao. “These represent the scenarios and challenges we have encountered in the analysis of liquid-phase electron microscopy videos.”
The researchers collected measurements from approximately 300,000 pairs of interacting nanoparticles.
As found in past studies by Chen’s group, contrast continues to be a problem while imaging certain types of nanoparticles. In their experimental work, the team used particles made out of gold, which are easy to see with an electron microscope. However, particles with lower molecular weights like proteins, plastic polymers and other organic nanoparticles show very low contrast when viewed under an electron beam.
“Biological applications, like the search for vaccines and drugs, underscore the urgency in our push to have our technique available for imaging biomolecules,“ Chen said. “There are critical nanoscale interactions between viruses and our immune systems, between the drugs and the immune system, and between the drug and the virus itself that must be understood. The fact that our new processing method allows us to extract information from samples as demonstrated here gets us ready for the next step of application and model systems.”
The team has made the source code for the machine learning program used in this study publicly available through the supplemental information section of the new paper. “We feel that making the code available to other researchers can benefit the whole nanomaterials research community,” Chen said.

Chen also is affiliated with the Chemistry Department, the Beckman Institute for Advanced Science and Technology and the Materials Research Laboratory at the University of Illinois.
The National Science Foundation and Air Force Office of Scientific Research supported this study.
Read the paper in full here.










