
ΑΙhub.org
Using machine learning to speed bioscaffold development
By Mike Williams
A team led by computer scientist Lydia Kavraki used a machine learning approach to predict the quality of scaffold materials produced by 3D-printing, given the printing parameters. The work also found that controlling print speed is critical in making high-quality implants.
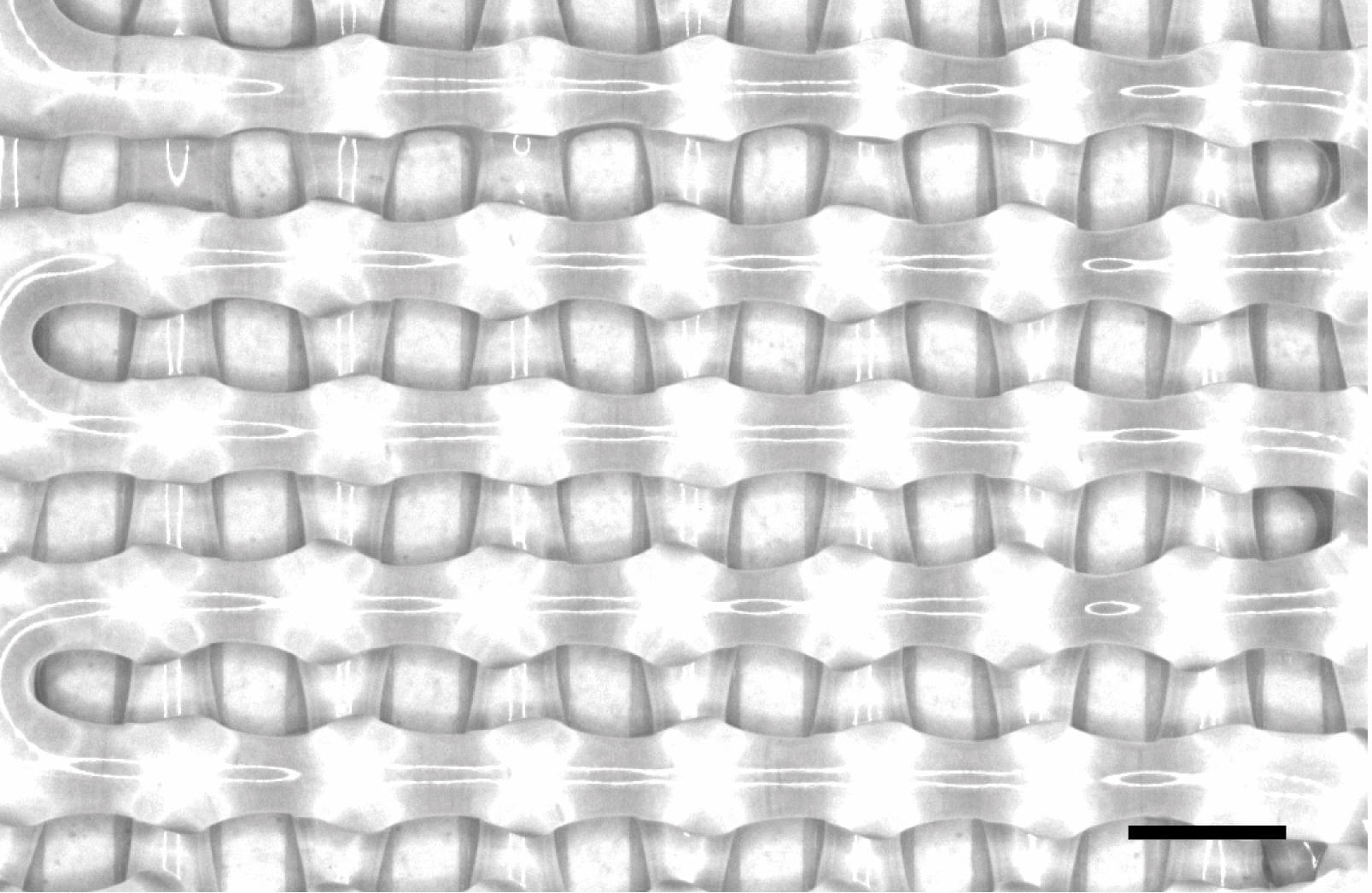
Bioscaffolds developed by co-author and bioengineer Antonios Mikos are bonelike structures that serve as placeholders for injured tissue. They are porous to support the growth of cells and blood vessels that turn into new tissue and ultimately replace the implant.
Mikos has been developing bioscaffolds to improve techniques to heal craniofacial and musculoskeletal wounds. That work has progressed to include sophisticated 3D printing that can make a biocompatible implant custom-fit to the site of a wound.
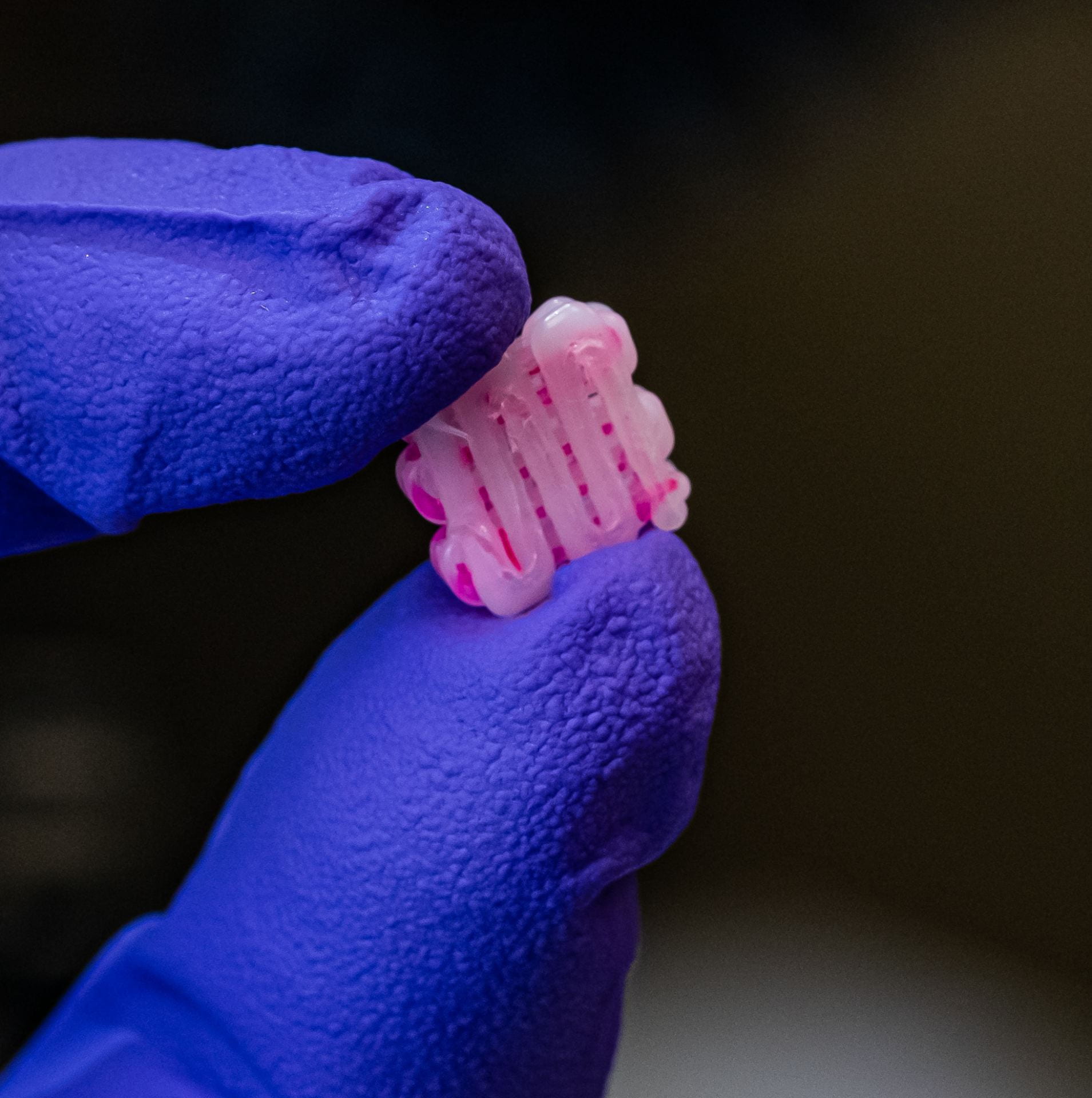
That doesn’t mean there isn’t room for improvement. With the help of machine learning techniques, designing materials and developing processes to create implants can be faster and eliminate much trial and error.
“We were able to give feedback on which parameters are most likely to affect the quality of printing, so when they continue their experimentation, they can focus on some parameters and ignore the others,” said Kavraki. The team reported its results in Tissue Engineering Part A.
The study identified print speed as the most important of five metrics the team measured, the others in descending order of importance being material composition, pressure, layering and spacing.
Mikos and his students had already considered bringing machine learning into the mix. The COVID-19 pandemic created a unique opportunity to pursue the project. “This was a way to make great progress while many students and faculty were unable to get to the lab,” Mikos said.
Kavraki said the researchers — graduate students Anja Conev and Eleni Litsa in her lab and graduate student Marissa Perez and postdoctoral fellow Mani Diba in the Mikos lab, all co-authors of the paper — took time at the start to establish an approach to a mass of data from a 2016 study on printing scaffolds with biodegradable poly(propylene fumarate), and then to work out what more was needed to train the computer models. “The students had to figure out how to talk to each other, and once they did, it was amazing how quickly they progressed,” Kavraki said.
The team explored two modeling approaches. One was a classification method that predicted whether a given set of parameters would produce a “low” or “high” quality scaffold. The other was a regression-based approach that approximated the values of print-quality metrics to come to a result. Kavraki said both relied upon a “classical supervised learning technique” called random forest that builds multiple “decision trees” and “merges” them together to get a more accurate and stable prediction.
Ultimately, the collaboration could lead to better ways to quickly print a customized jawbone, kneecap or bit of cartilage on demand.
“A hugely important aspect is the potential to discover new things,” Mikos said. “This line of research gives us not only the ability to optimize a system for which we have a number of variables — which is very important — but also the possibility to discover something totally new and unexpected. In my opinion, that’s the real beauty of this work.”
“It’s a great example of convergence,” he said. “We have a lot to learn from advances in computer science and artificial intelligence, and this study is a perfect example of how they will help us become more efficient.”
“In the long run, labs should be able to understand which of their materials can give them different kinds of printed scaffolds, and in the very long run, even predict results for materials they have not tried,” Kavraki said. “We don’t have enough data to do that right now, but at some point we think we should be able to generate such models.”
About the authors
 Lydia Kavraki is the Noah Harding Professor of Computer Science and a professor of bioengineering, mechanical engineering and electrical and computer engineering at Rice University.
Lydia Kavraki is the Noah Harding Professor of Computer Science and a professor of bioengineering, mechanical engineering and electrical and computer engineering at Rice University.
 Antonios Mikos is the Louis Calder Professor of Bioengineering and Chemical and Biomolecular Engineering and a professor of chemistry and materials science and nanoengineering at Rice University.
Antonios Mikos is the Louis Calder Professor of Bioengineering and Chemical and Biomolecular Engineering and a professor of chemistry and materials science and nanoengineering at Rice University.









