
ΑΙhub.org
#NeurIPS2020 invited talks round-up: part three – causal learning and the genomic bottleneck

In this post we conclude our summaries of the NeurIPS invited talks from the 2020 meeting. In this final instalment, we cover the talks by Marloes Maathuis (ETH Zurich) and Anthony M Zador (Cold Spring Harbor Laboratory).
Marloes Maathuis: Causal learning
Marloes began her talk on causal learning with a simple example of the phenomenon known as Simpson’s paradox, in which a trend appears in several different groups of data but disappears or reverses when these groups are combined. She also talked about the importance of considering causality when making decisions based on such data.
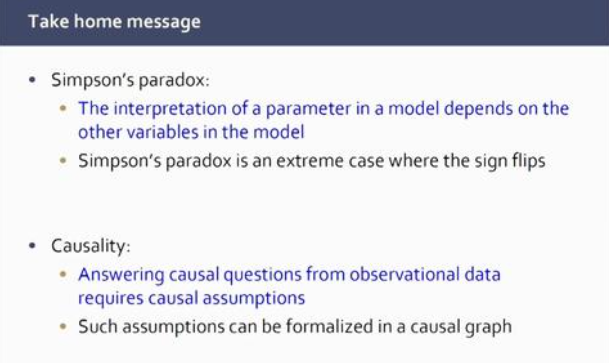
Marloes went on to explain the difference between causal and non-causal questions. Non-causal questions are about predictions in the same system, for example, predicting the cancer rate among smokers. Causal questions, on the other hand, are about the mechanism behind the data or about predictions after an intervention to the system. For example, asking if smoking causes lung cancer, or predicting the spread of a virus epidemic after imposing new regulations.
Causal questions are ideally answered by randomised controlled experiments. However, sometimes it is not possible to carry out these experiments, so we need to estimate causal effects from observational data. Marloes described her research into determining methodology, using causal directed acyclic graphs (DAGs) to estimate such causal effects.
In the final part of her presentation, Marloes explained the methodology used when the causal graph is unknown. One possible approach is to hypothesize possible DAGs. Another approach is to learn the DAG from the data.
To find out more you can watch the talk in full here.
Anthony M Zador: The genomic bottleneck: a lesson from biology
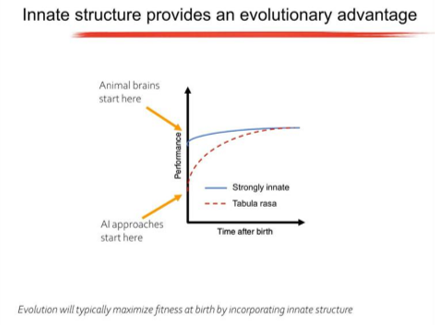
Anthony spoke about the innate abilities that animals have and argued that most animal behaviour is not the result of clever learning algorithms, but is encoded in the genome. Specifically, animals are born with highly structured brain connectivity, which enables them to learn very rapidly. Examples of innate ability include birds making species-specific nests and beavers building dams. Having these abilities as innate provides an evolutionary advantage.
In the talk, Anthony outlined the number of parameters it takes to wire the brains of different creatures. For C elegans (a type of worm), the simplest animal studied, 302 neurons with 7000 synapses are needed. The genome of C elegans consists of about 200 million bits (where two bits make a nucleotide). These 200 million bits are easily enough to specify the precise wiring of 7000 synapses.
Compare this to a human brain: we have roughly 1011 neurons and 1014 synapses. It is estimated that it takes about 1015 bits to specify a human brain. However, our genome is only 109 bits. Anthony explained this missing factor of 106: the genome doesn’t specify every single synapse, rather, it specifies rules for wiring up the brain.
This led onto discussion of the notion that the wiring diagram needs to be compressed through a “genomic bottleneck”. The genomic bottleneck suggests a path toward AI architectures capable of rapid learning and in the final part of his talk, Anthony outlined some of the research that he is carrying out in this area.
Watch the talk here.
tags: NeurIPS, NeurIPS2020










