
ΑΙhub.org
DeepMind and EMBL release database of predicted protein structures
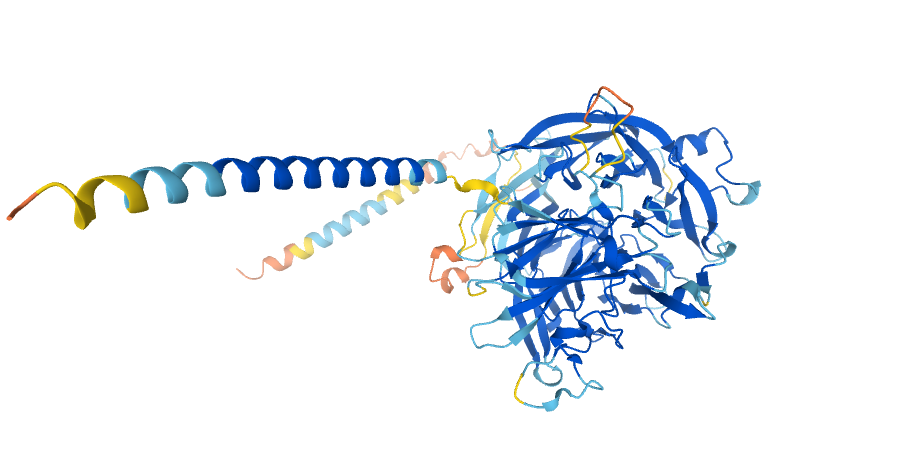
T-cell immunomodulatory protein homolog, from the AlphaFold Protein Structure Database, reproduced under a CC-BY-4.0 license.
DeepMind and the European Molecular Biology Laboratory (EMBL) have partnered to produce a database of predicted protein structure models.
The first release covers all ~20,000 proteins expressed in the human proteome, and the proteomes of 20 other biologically significant organisms, totalling over 350k structures. In the coming months they plan to expand the database to cover a large proportion of all catalogued proteins (the over 100 million in UniRef90).
The data is freely and openly available to the scientific community. You can access the AlphaFold Protein Structure Database here.
Back in November, DeepMind reported on their AlphaFold system that was able to predict, with high accuracy, a protein’s 3D structure from its amino acid sequence. We wrote about it here. This database is the next step in the journey, and the collaborators hope that this will be a useful tool for researchers and open up new avenues for scientific discovery.
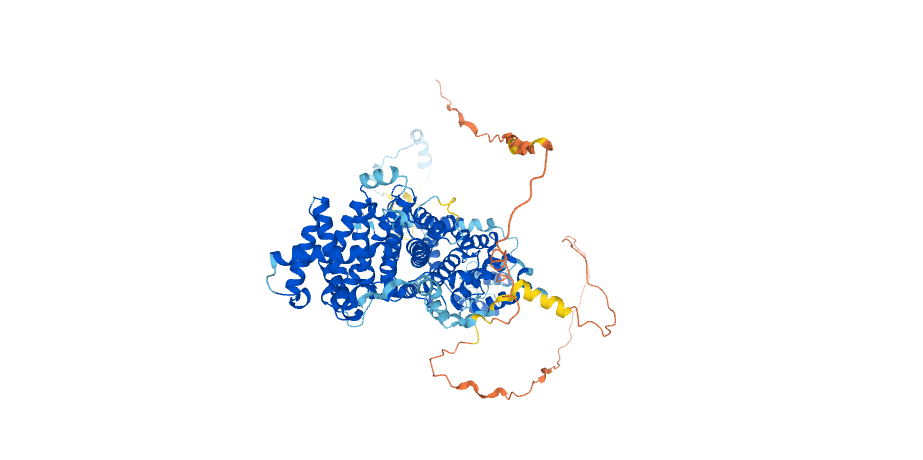
Another example protein structure from the AlphaFold Protein Structure Database, reproduced under a CC-BY-4.0 license. This is Striatin-interacting protein 1. It plays a role in the regulation of cell morphology and cytoskeletal organization, required in the cortical actin filament dynamics and cell shape. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. The parts of the protein with a pLDDT score of above 90 are shown in dark blue, between 70 and 90 in light blue, between 50 and 70 in yellow, and below 50 in red.
In a recently published Nature article, Highly accurate protein structure prediction with AlphaFold, you can find out more about the neural network-based model and methodology that the AlphaFold team used. In this second Nature article, Highly accurate protein structure prediction for the human proteome, published yesterday, you can read more about the application of AlphaFold to the human proteome.
Find out more
AlphaFold Protein Structure Database
DeepMind blog post
EMBL-EBI news article
Highly accurate protein structure prediction with AlphaFold, Nature article.
Highly accurate protein structure prediction for the human proteome, Nature article.
DeepMind open source code
AlphaFold Colab
tags: quick read










