
ΑΙhub.org
#IJCAI2022 invited talk: Insights in medicine with Mihaela van der Schaar

The 31st International Joint Conference on Artificial Intelligence and the 25th European Conference on Artificial Intelligence (IJACI-ECAI 2022) took place from 23-29 July, in Vienna. As part of the conference there were eight fascinating invited talks. In this post, we summarise the presentation by Mihaela van der Schaar, University of Cambridge. The title of her talk was “Panning for insights in medicine and beyond: New frontiers in machine learning interpretability”.
Mihaela began by explaining why the field of medicine is so complex. Differences between individuals, due to factors such as genetic background, environmental exposure, and life-style, lead to variations in symptoms, disease trajectories, and responses to treatments. What clinicians have to do is make judgments about each patient based on this very complex web of information. The goal of Mihaela’s research lab is to develop machine learning methods which both address complex problems in medicine, and impower clinicians and patients.
Mihaela believes that there are many opportunities for using machine learning in medicine. For example, it could be used to enable precision medicine, to help understand disease trajectories, to help inform and improve clinical pathways, and to aid new discoveries, to name a few. Mihaela and her team work closely with clinicians to try to understand and model complex problems in medicine. They are developing human-machine partnerships where machine learning augments human skills.
 Mihaela delivering her talk at IJCAI-ECAI2022. The opportunities for machine learning in medicine.
Mihaela delivering her talk at IJCAI-ECAI2022. The opportunities for machine learning in medicine.
To highlight this close collaboration with clinicians, Mihaela pointed to engagement sessions that she initiated two years ago, and so far have involved around 500 clinicians. In these sessions, the clinicians discuss what type of tools they’d like to have and they provide input on the design. If you’d like to watch these sessions they have been recorded and are available on YouTube (see an example here).
The ability to provide explanations is very important in the field of medicine. Mihaela and her team asked clinicians what they wanted from an explanation. They identified three different areas: 1) explanatory patient features and examples, and the ability to explain time-series data and treatment trajectories, 2) personalised explanations – rather than a “one size fits all” explanation, this would provide clinicians with explanations based on specific patients that they select, 3) discovery of governing equations of medicine.
With regards to the third area – the discovery of governing equations using machine learning – Mihaela explained that what she has in mind here is learning equations from data. These could be explicit functions, implicit functions, or ordinary differential equations (ODEs). She believes that we will only have concise, generalizable, transparent and interpretable methods once we’ve been able to distil the laws of medicine into governing equations.
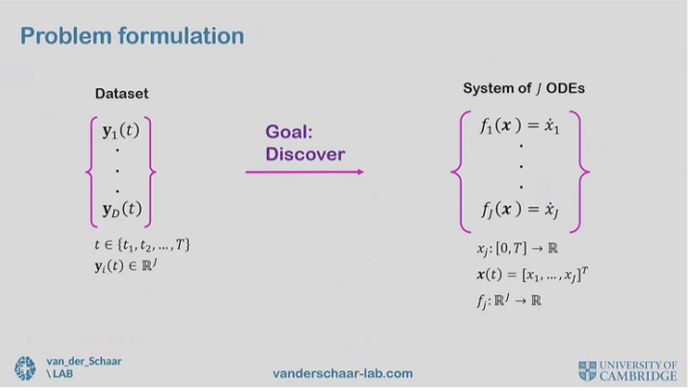 Formulating the problem of using data to discover underlying ODEs. Screenshot from Mihaela’s talk.
Formulating the problem of using data to discover underlying ODEs. Screenshot from Mihaela’s talk.
The most exciting and challenging frontier of these is learning ODEs. After all, the human body is a dynamical system and we’d like to understand how it changes over time. The problem formulation as follows: there is a dataset of trajectories, and we’d like to learn underlying differential equations.
In recent work, Mihaela and co-authors proposed the Discovery of Closed-form ODE framework (D-CODE). The key insight behind D-CODE is the variational formulation of ODE, which establishes a direct link between the trajectory ![]() (t) and the ODE f while bypassing the unobservable time derivative
(t) and the ODE f while bypassing the unobservable time derivative ![]() (t). They developed a novel objective function based on this insight, and proved that it is a valid proxy for the estimation error of the true (but unknown) ODE.
(t). They developed a novel objective function based on this insight, and proved that it is a valid proxy for the estimation error of the true (but unknown) ODE.
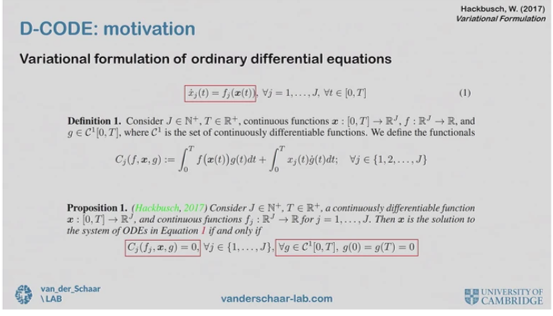 D-CODE. Screenshot from Mihaela’s talk.
D-CODE. Screenshot from Mihaela’s talk.
In the experiments, D-CODE successfully discovered the governing equations of a diverse range of dynamical systems under challenging measurement settings with high noise and infrequent sampling. One experiment in particular concerned learning an equation that governs the growth of tumours and the impact of chemotherapy. They used data from eight clinical trials that followed patients over time, and found that their method provided an equation that gave a much better representation of data than previous methodologies.
If you are interested in finding out more about Mihaela’s work, here are some links that she highlighted during her talk:
Engagement sessions with clinicians
Inspiration exchange engagement sessions
D-CODE: Discovering Closed-from ODEs from observed trajectories
tags: IJCAI2022











