
ΑΙhub.org
Few-shot learning for medical image analysis
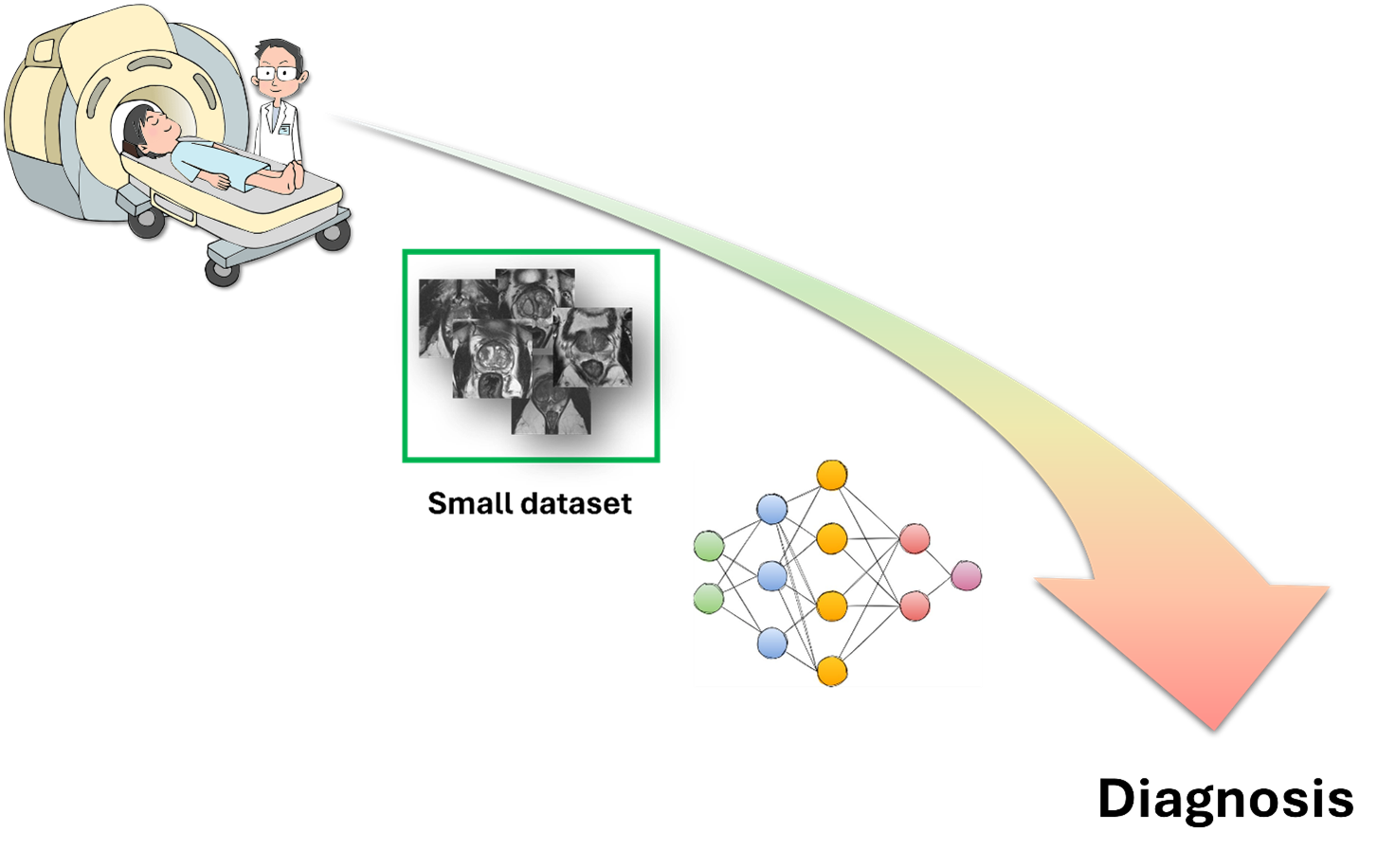
Is few-shot learning the gateway to integrating AI into medicine for good? Let’s explore the current state of the art.
Not too long ago, the concept of Artificial Intelligence (AI) resided primarily within the realm of academics and the fantastical landscapes of science fiction movies, often linked to the idea of humanoid robots. In recent years, the advancement in computing power and speed has catalyzed an unprecedented surge in AI development, making it part of our daily lives. But it doesn’t stop here. With the advent of innovative technologies such as text-to-image models like Dall-E and chatbot-type models like ChatGPT, AI is now part of our everyday jargon. Indeed, these breakthroughs have democratized AI, allowing even those with basic computer knowledge to harness its power. It’s no wonder that AI has become a topic on everyone’s lips.
Apart from creating images from text or chatting with humans like a pro, AI also has the potential to make great strides in the medical field, like performing non-invasive diagnoses or detecting diseases from images. However, most of these applications are still stuck in research labs, and only a few are employed in clinical practice. The main reason why AI can’t do as much as it does in other fields is that it doesn’t have enough data to learn from. Unlike humans, who can learn a task with just a few examples and apply it to different situations, AI models need tons of data to learn all the patterns and generalize well.
 Deep learning is data hungry.
Deep learning is data hungry.
To overcome this limitation, researchers have come up with different ways to train models to learn the most important features of a given task and adapt quickly to new ones. One of these ways is called few-shot learning (FSL). As the name suggests, FSL means learning from just a few examples. But how can a model learn from so little data? Well, one trick is to use meta-learning. Meta-learning means learning how to learn. Instead of training a model on one task, such as dogs vs cats, you train it on many related tasks, such as birds vs fish, horses vs cows, etc. Then, you use another algorithm to update the model based on how well it performs on these tasks. This way, the model learns not only the features of each task but also the best way to learn new tasks so that it can quickly adjust its parameters and make accurate predictions.
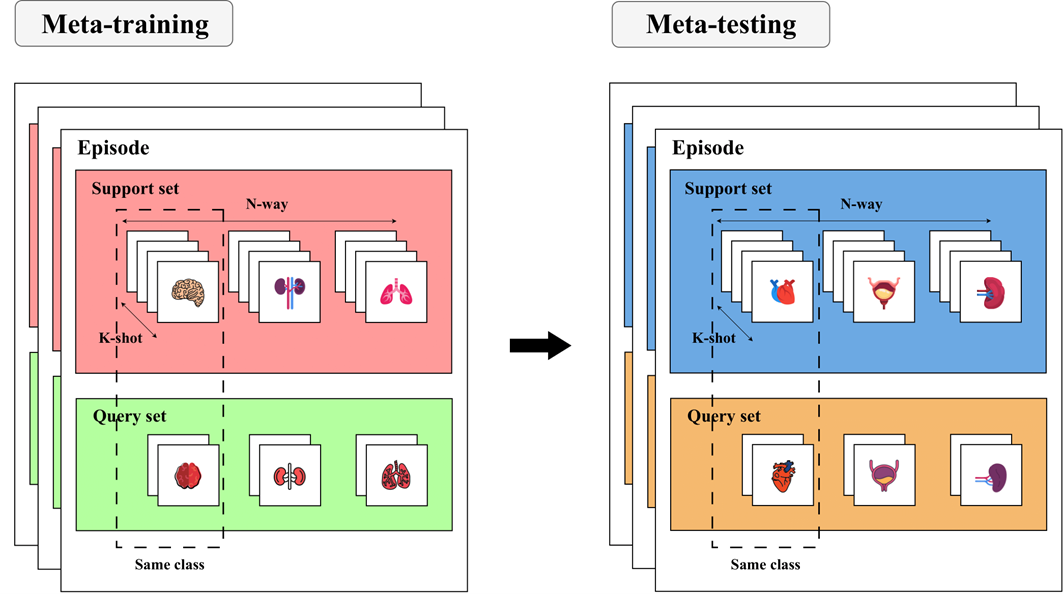 The most popular way to perform meta-learning is through the N-way K-shot paradigm.
The most popular way to perform meta-learning is through the N-way K-shot paradigm.
In our work [1], we wanted to investigate the state of the art of FSL in medical imaging since we believe that such methods can promote the use of AI in clinical practice, providing a series of advantages that we’ll describe below. Medical datasets are usually small because images are hard to get and label because of privacy issues, high costs, and expert work. FSL could overcome these problems since it requires only a little data. In addition, FSL could deal with rare medical conditions that don’t have enough data for classical deep-learning methods by exploiting information learned from common diseases to adapt to new and rare cases with few examples. Last but not least, FSL has potential in personalized medicine, where models have to analyze images from individual patients. With just a few examples from each patient, FSL models could adjust their analysis based on the patient’s specific features, improving the accuracy and quality of medical diagnosis and treatment.
We examined about 80 articles, grouping them based on the primary outcomes: segmentation, classification, and registration. Our focus was on meta-learning as the key framework for addressing FSL. For each article, we highlighted whether meta-learning was employed and, if so, what type. Additionally, we outlined the anatomical district(s) investigated and reported the best performance achieved in each case. We also conducted a thorough analysis of the risk of bias and applicability for each paper to inform the reader of the real impact of the work. To offer a comprehensive overview, we finally defined a high-level pipeline that incorporates the methods from all the reviewed works.
We found out that most papers (61%) used FSL for medical imaging for segmentation and that different anatomical districts got different attention and results. Concerning segmentation and registration tasks, the heart was the most investigated organ, while in the classification domain, most papers focused on lung images. Examining the numerical results, the most successful segmentation tasks involved identifying the aorta and left ventricle. However, we found that FSL techniques still struggle in segmenting the prostate. Speaking of classification, FSL provides compelling results in distinguishing skin lesions and eye diseases. Conversely, its performance lags in the case of liver and lung diseases. Furthermore, FSL exhibited promising potential in registering heart images, indicating a positive direction for further research and development.
Despite the success of meta-learning for FSL, only half of segmentation and classification tasks actually used meta-learning methods, while none of the registration studies used them. Among the subset of works that employed meta-learning, it’s noteworthy that metric-learning-based methods were the most prevalent choice, followed by initialization-based and hallucination-based methods. In terms of performance, non-meta-learning methods consistently outshone their meta-learning counterparts across all three outcomes, even though hallucination-based methods showed promising results.
In terms of robustness evaluation, we observed that several studies, particularly those in classification and registration tasks, did not conduct comprehensive model investigation analyses. This gap can potentially lead to incomplete and unreliable performance assessments. In addition, we noted issues related to the risk of bias in some segmentation and classification studies, which lacked clarity in explaining how they addressed the FSL task and the number of training data employed.
Our final contribution involved creating a comprehensive pipeline based on the synthesis of all the reviewed work. We outlined three core stages: pre-training, fine-tuning, and data augmentation. We declined each step into subgroups of training and data augmentation methods, as depicted in the following figure.
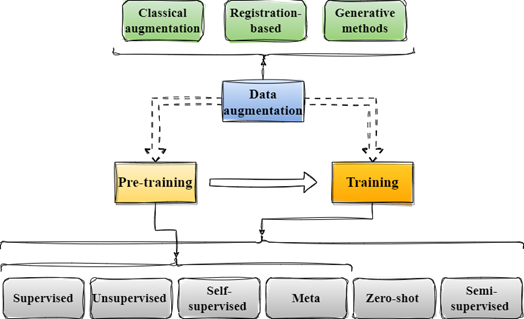 General pipeline for the methods used by the reviewed papers.
General pipeline for the methods used by the reviewed papers.
For each article, we documented the specific subcategories utilized. Notably, we found that immediately after meta-learning, the prevailing technique for training is classical supervised learning, often customized through new modules designed to enhance the model’s performance and generalization abilities. Furthermore, we noticed that in segmentation studies, there was a common trend of incorporating semi-supervised learning techniques, where labelled and unlabeled data are simultaneously used to refine the segmentation process.
In light of these findings, we encourage future researchers in the field to consider the following actions:
- Delve into less explored meta-learning methods, such as hallucination-based approaches, which showed promising results;
- Invest in creating new methods for less-performing applications, such as prostate segmentation and lung or liver diseases classification;
- Prioritize thorough model validation and comprehensive analyses to facilitate fair comparisons and the practical implementation of FSL models in clinical settings.
Our systematic review is intended to serve as a valuable resource for future researchers in the field, offering guidance on areas of anatomical interest and methodological exploration that warrant further investigation. Our goal is to promote the advancement and broader adoption of FSL techniques within the medical imaging domain by addressing identified gaps, emphasizing robustness evaluation and showing an overview of the methods currently used in the state of the art.
References
[1] Pachetti, E., & Colantonio, S. (2023). A Systematic Review of Few-Shot Learning in Medical Imaging. arXiv preprint.
Icons downloaded from www.flaticon.com.












