
ΑΙhub.org
Machine learning enhances chemical analysis at the nanoscale
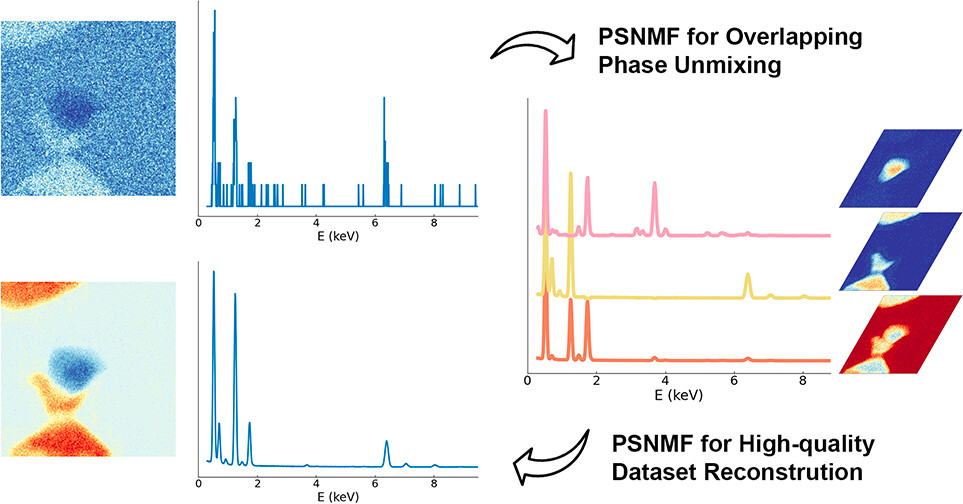 Introducing a non-negative matrix factorization based pan-sharpening (PSNMF) method to determine chemical compositions from noisy x-ray spectroscopy data. From: Nano Lett. 2024, 24, 33, 10177-10185. Authors: Hui Chen, Duncan T. L. Alexander, Cécile Hébert.
Introducing a non-negative matrix factorization based pan-sharpening (PSNMF) method to determine chemical compositions from noisy x-ray spectroscopy data. From: Nano Lett. 2024, 24, 33, 10177-10185. Authors: Hui Chen, Duncan T. L. Alexander, Cécile Hébert.
By Nik Papageorgiou
“Nanomaterials” is a broad term used to describe chemical substances or materials in which a single unit is sized between 1 and 100 nanometers (a nanometer is a billionth of a meter). They include exotic materials such as carbon nanotubes, silver nanoparticles (used as antimicrobials), nanoporous materials, and many types of catalysts used for efficiently driving chemical reactions.
Nanomaterials are currently used in a wide range of fields, from medicine to electronics, which means that the ability to determine their exact chemical composition is essential. Nonetheless, this proves challenging, because traditional methods for analyzing nanomaterials tend to be susceptible to low signal-to-noise ratios.
For example, one extensively used method is energy-dispersive X-ray spectroscopy (EDX), combined with scanning transmission electron microscopy. This technique provides detailed maps of where different elements are located within a sample, but it often produces noisy data, especially on such small objects, and mixed signals when different materials overlap, making precise chemical analysis difficult.
The noisy data are usually “cleaned up” with various techniques, from simple spatial filtering to more sophisticated machine learning approaches like principal component analysis, that separate the signals from the noise, but they too have their drawbacks. For example, they can introduce errors, or struggle to distinguish between chemical signals when they are very similar.
Now, three scientists at EPFL, Hui Chen, Duncan Alexander, and Cécile Hébert have developed a machine learning-based method called PSNMF (“non-negative matrix factorization-based pan-sharpening”) that enhances the clarity and accuracy of EDX data, making it easier to identify and quantify different chemical elements in nanomaterials.
The team started by leveraging a special characteristic of their data called “Poisson noise”. This type of noise occurs because the detection of X-ray photons is random. When the electron beam hits the sample, it produces X-ray photons, but the number detected varies randomly each time, creating a noisy, grainy pattern known as Poisson noise.
To improve the clarity of their data, the researchers combined data from nearby pixels, enhancing the signal-to-noise ratio in the spectrum at the cost of the spatial resolution.
They then applied a machine learning method called “non-negative matrix factorization” (NMF) to this clearer dataset. NMF is a mathematical technique that breaks down a large dataset into simpler, smaller parts, ensuring all parts are non-negative, which helps identify patterns in the data. This approach gave them good spectral data at the cost of having blurry images with large pixels.
Next, they repeated the NMF process on the original high-resolution dataset to preserve detailed spatial information, but initializing the factorization with the previously identified spectral components. Finally, they combined the results from both steps to produce a high-quality dataset, that has both high spectral fidelity and high spatial resolution.
The researchers validated PSNMF using synthetic data, computed thanks to a modelling algorithm developed in the lab. Those data mimicked real-world challenges, such as analyzing mineral samples formed under extreme conditions. The method proved highly effective, accurately identifying and separating different materials, even those in tiny amounts.
When applied to actual samples, including a nanomineral and a nanocatalyst, PSNMF successfully separated and quantified overlapping materials. This precise analysis is crucial for understanding and developing new technologies that rely on these complex nanostructures.
PSNMF is a significant improvement in nanoscale chemical analysis. By providing accurate results despite noisy data and overlapping signals, this method enhances our ability to study and utilize nanomaterials in various fields, from advanced electronics to medical devices.
Read the work in full
Leveraging Machine Learning for Advanced Nanoscale X-ray Analysis: Unmixing Multicomponent Signals and Enhancing Chemical Quantification, Hui Chen, Duncan T. L. Alexander, Cécile Hébert, Nano Letters (2024).









