
ΑΙhub.org
AI in cancer research & care: perspectives of three KU Leuven institutes

In 2021, cancer was the second leading cause of death in the European Union. Notably, while Europe constitutes only a tenth of the global population, it accounts for almost a quarter of the world’s cancer cases, bearing an economic impact of approximately €100 billion annually. Belgian statistics further highlight that every step forward in cancer treatment and care could significantly alleviate the immense personal and societal burden.
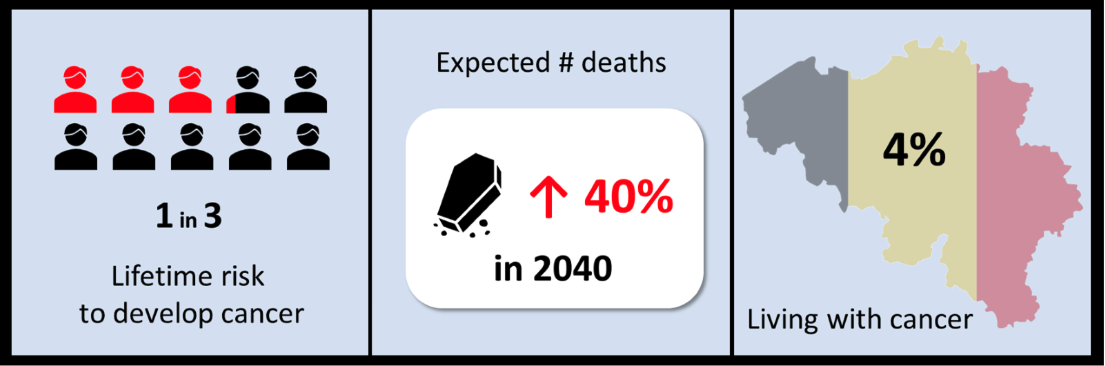 Figure 1: Belgian cancer statistics. Belgian cancer registry
Figure 1: Belgian cancer statistics. Belgian cancer registry
Globally, extensive efforts are made through a variety of innovative approaches with the ultimate objectives of better prevention, earlier detection, and improved patient outcomes and care. Personalized medicine is considered the holy grail of cancer care in most of these initiatives. Tailoring interventions to the unique characteristics of individual patients promises to revolutionize cancer care. Artificial intelligence (AI) plays a pivotal role in this transformative journey towards precision medicine, aiding researchers and healthcare professionals in accurately predicting cancer risks, enabling earlier diagnoses, and customizing treatment plans to meet individual needs. This increases the likelihood of successful outcomes while at the same time minimizing drug resistance and adverse effects.
Beyond ensuring optimal treatment and care, personalized or precision medicine will be crucial for sustainable healthcare and the efficient use of social security resources. A notable instance is immunotherapy, where approximately 30% of the 40,000 Flemings diagnosed with cancer annually receive a form of this treatment, costing on average €150,000 per patient. This translates to an estimated €2.2 billion in expenses, with effectiveness observed in only half of these patients. Consequently, about €1.1 billion is spent suboptimally each year on immunotherapy alone. Enhancing diagnostics and delivering tailored, cost-efficient treatments are therefore essential to maintaining the financial health of our social security system.
A pivotal move towards achieving this goal involves the development of new, often disruptive technologies that grant access to extensive information about cancer patients and their tumor biology, albeit often hidden within large datasets. Fully leveraging this data will be key to advancing fundamental and translational cancer research, and finally unlocking the full potential of personalized medicine. AI, in this regard, has proven to be a formidable tool for identifying patterns and predicting outcomes from complex datasets, developing accurate prognostic models, and integrating diverse data sets, thereby accelerating the progress in personalized medicine.
This story is a collaboration of three KU Leuven Institutes that are working at the intersections of cell research, cancer research and care, and artificial intelligence across KU Leuven and UZ Leuven:
- KU Leuven Institute for Single Cell Omics (LISCO) unites researchers with expertise in the development of single-cell and spatial multi-omics technologies and their application in various (biomedical) domains including oncology.
- Leuven Cancer Institute (LKI) unites fundamental, translational, and clinical researchers alongside physicians, nurses and allied health professionals, all dedicated to delivering optimal cancer patient care.
- Leuven.AI is fostering fundamental and applied AI research. A long-standing research focus is on biomedical applications of AI.
Medical imaging
From an X-ray scan of your teeth to an ultrasound scan of a foetus during pregnancy, most of us have already had a medical imaging examination. It enables us to examine the interior of our bodies, and although it’s one of the oldest methods for diagnosing cancer, medical imaging remains a game-changer when it comes to diagnosing, planning treatment, and monitoring cancer patients. In the early days of medical imaging, X-rays and CT scans were the only means of peering inside a patient’s body. However, we now have many imaging techniques at our disposal to explore beyond what is immediately visible. It enables us to visualize and locate different structures and masses within the body and provides insights into factors such as metabolic activity or vascularity of a tumor.
 Figure 2: Radiation therapy in the Leuven proton therapy center, copyright UZ Leuven.
Figure 2: Radiation therapy in the Leuven proton therapy center, copyright UZ Leuven.
By tapping into the power of different imaging methods such as MRI (magnetic resonance imaging), PET (positron emission tomography) and SPECT (single-photon emission computer tomography) scans, clinicians gain a deeper understanding of the tumor complexity. This enables them to make more educated and hence more accurate decisions when it comes to diagnosis, identification of a (personalized) treatment plan, monitoring of treatment efficacy and long-term relapse surveillance. Innovation in the medical imaging field has resulted in images with increasing sensitivity and specificity, allowing more true-positive detection of lesions and more false-positive detections to be excluded. Yet, optimal analysis and interpretation of these results remain crucial to make use of its full potential. Such accurate interpretation of medical images requires expert knowledge and an extremely trained eye to identify subtle abnormalities. Given the increasing volumes and complexity of medical imaging data, properly retrieving the vast amount of information comes with a significant time investment. And these are exactly the challenges where AI can provide a huge added value. While the different medical imaging modalities differ in terms of underlying technology, visual representation or medical purpose, from the viewpoint of AI they all come down to a combination of pixels, waiting to be analyzed.
Before the emergence of deep learning, AI-driven medical imaging, or computer vision-based methods, depended on meticulously designed features like color, shape, texture, and their combinations. These features tried to capture the particularities of a tumor for an algorithm being able to find it. This process required that these ‘traditional’ features be manually created by medical experts, which was often a time-consuming task.
The introduction of deep learning significantly advanced medical imaging by eliminating the need to manually design target features. Instead, these newer methods, utilizing cascades of artificial neuron layers, could autonomously generate and identify relevant features (Fig. 3). It enabled the detection of minor features such as edges, corners, or blobs, from image areas spanning just a few pixels. As we move up layers in the neural architecture, these basic features would merge into higher-level features across broader sections of the images. Consequently, these techniques could recognize complex shapes and patterns associated with tumors, like the edges of a tumor’s boundary or biomarkers indicating the state of the cancer or the response to treatment. Algorithms based on deep learning for spotting and classifying these patterns and anomalies allow for quicker and more precise detection in extensive datasets of high-resolution images than what the human eye can achieve.
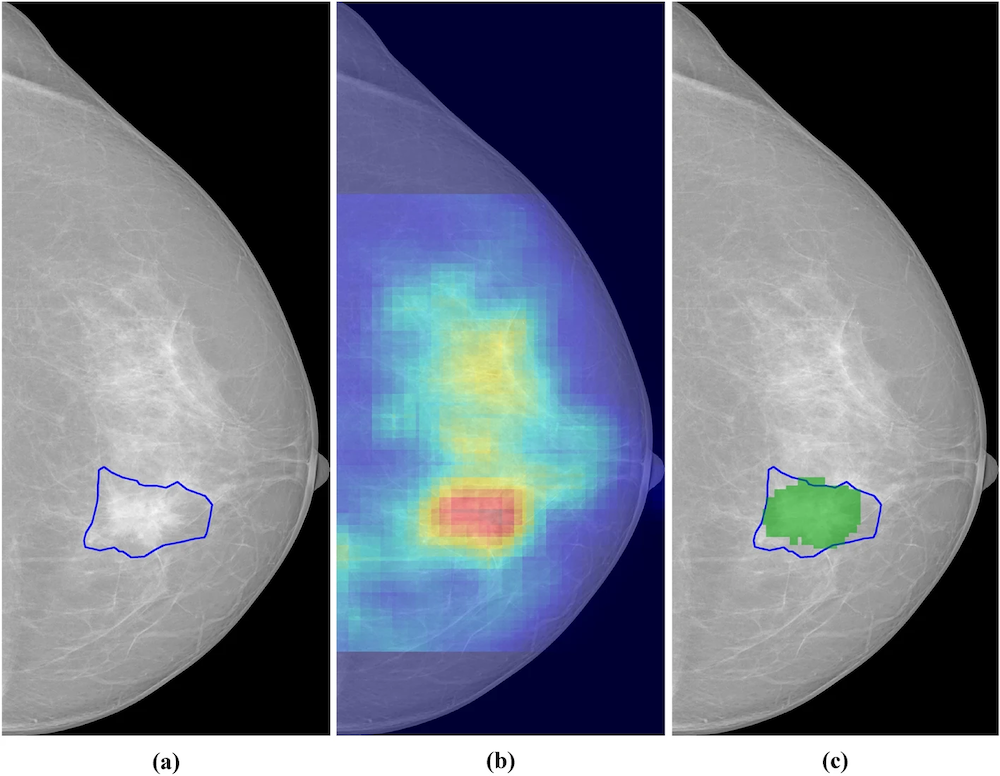 Figure 3: Segmentation of breast cancer lesions. (a) Manually segmented lesion,(b) Saliency map for an AI system, (c) Relevant region (in green) obtained by thresholding the saliency map in (b). Reprinted under the Creative Commons Attribution 4.0 International License from DOI:10.1038/s41598-023-46921-3
Figure 3: Segmentation of breast cancer lesions. (a) Manually segmented lesion,(b) Saliency map for an AI system, (c) Relevant region (in green) obtained by thresholding the saliency map in (b). Reprinted under the Creative Commons Attribution 4.0 International License from DOI:10.1038/s41598-023-46921-3
Such (partly) automated processes in object detection and image segmentation support healthcare professionals in surgical planning, outlining the area for radiation, calculating radiation dose distributions (Fig. 4), and monitoring tumor progression over time. Furthermore, the adoption of automated and optimized workflows, where repetitive tasks like data extraction, labeling, and standardization no longer solely depend on manual efforts, gives radiologists, and medical practitioners working with images in general, more time to concentrate on determining the best patient care trajectory.
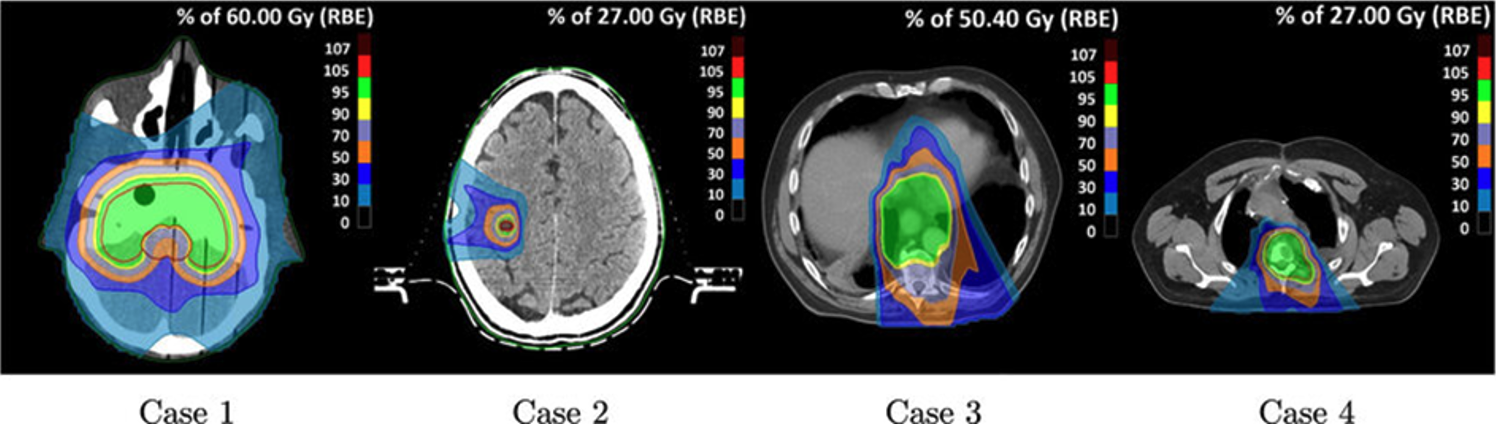 Figure 4: Axial slices of one phantom (case 1) and three clinical (case 2-4) cases with overlaid estimated radiation dose distributions, based on dose calculations and plan optimization. DOI:10.1088/1361-6560/ac8dde
Figure 4: Axial slices of one phantom (case 1) and three clinical (case 2-4) cases with overlaid estimated radiation dose distributions, based on dose calculations and plan optimization. DOI:10.1088/1361-6560/ac8dde
Implementing AI algorithms at various stages of the image analysis process has proven to be beneficial for healthcare professionals in several ways. For instance, the application of deep learning models has led to a 21% improvement in the sensitivity of detecting brain metastases. This reinforces the notion that images hold information beyond what traditional analyses can extract. This was further supported by the finding that a hybrid model, combining AI-based and radiologist predictions, reduced false-positive breast cancer findings in ultrasound images by 37.3%, subsequently lowering the necessity for patient biopsies. Unlocking the full anatomical and functional capacity of whole-body diffusion-weighted MRI is also the goal of KU and UZ Leuven researcher and radiologist Prof. Vincent Vandecaveye. Developing AI tools for segmentation and feature identification in these MRI images will enable him and his team to enhance the diagnosis, metastasis detection, and therapy monitoring across various tumor types.
Another significant advantage of AI-based detection is the marked reduction in both inter- and intra-observer variability among clinicians, which leads to less subjectivity in identifying and interpreting abnormalities and, therefore, in making treatment plan decisions. This benefit is exemplified in an interdisciplinary study on head and neck cancer led by Prof. Frederik Maes and Prof. Sandra Nuyts at KU and UZ Leuven. In addition, colleague Prof. Wouter Crijns is involved in integrating these AI-contouring and treatment planning tools into radiotherapy guidelines, showcasing the practical application and impact of AI in reducing variability and improving treatment accuracy. This technology allows for the prediction of treatment plans containing up to 40 000 treatment unit parameter settings.
”Currently, AI image processing is assisting in existing tasks of the radiotherapy treatment preparation in daily clinical practice. But, most interestingly, the use of AI increases the receptive field of our image processing tools. In the near future, we will be able to analyze multiple images at once resulting in both multi-modal and longitudinal tools that surpass the abilities of current radiotherapy treatment preparation.”
— Prof. Wouter Crijns
Single-cell and spatial multi-omics
Our bodies are composed of trillions of cells, each containing roughly the same genetic material, or DNA. As you might recall from high school biology, this DNA is formed from long strands comprising four different chemicals, represented as A, T, C, and G. These chemicals carry the instructions for a cell’s structure, function, and behavior. To activate a specific instruction, a segment of the DNA is transcribed into RNA, a shorter messenger molecule that uses a similar four-chemical code. The proteins, which are the actual workhorses of the cell, are then produced based on these RNA molecules (Fig. 5). In cancer cells, alterations in this genetic information (or mutations) have accumulated during life, affecting the instructions. This changes the resulting molecules and the cell’s behavior and function, leading the cell to multiply uncontrollably, spread to other parts of the body, and sometimes resist treatment. In essence, the root cause of cancer can be traced to molecular alterations within our cells.
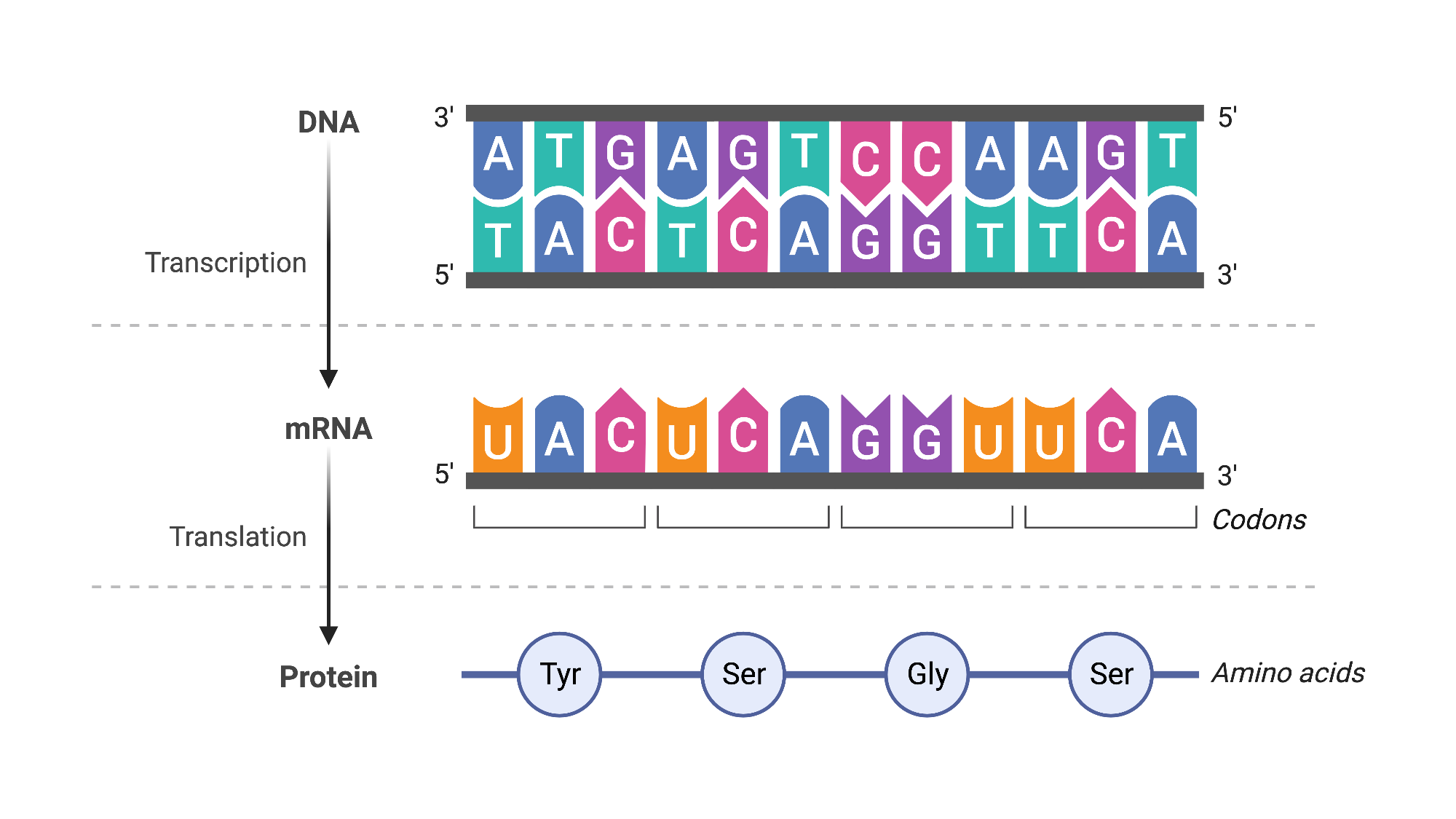 Figure 5: Central dogma of molecular biology. BioRender (2019).
Figure 5: Central dogma of molecular biology. BioRender (2019).
Single-cell omics: back to the basics
Single-cell omics refers to a set of methods and technologies that allows us to study these molecular anomalies in cancer cells. Specifically, each omics approach is designed to analyze a distinct class of biomolecules within individual cells. Genomics, for example, determines the DNA sequence within cells, transcriptomics characterizes and quantifies RNA, and proteomics deals with proteins. Through these technologies, we can identify mutations in the DNA or understand which specific cell functions have been altered. In fundamental research, this detailed molecular information helps us to learn what goes wrong in a tumor at the molecular level, guiding the development of new therapies or explaining why a tumor might not respond to a certain treatment. Additionally, these technologies can be applied clinically to identify and characterize cancer cells in liquid biopsies from patients, supporting non-invasive cancer diagnosis, treatment selection, and follow-up.
Despite their vast potential, single-cell omics technologies also pose challenges, particularly in the volume of data they generate and the complexity of analyzing this data. For instance, when employing short-read sequencing to decipher the chemical A,C,T,G-composition of DNA or RNA, the output for an individual cell can range from tens of thousands to several millions of fragments, each dozens of letters long. These fragments are then compared and mapped to a reference sequence (Fig. 6). The number of cells analyzed can vary widely, from a few to hundreds of thousands, depending on the aims and design of the specific study. Given the immense size of the dataset, which can be in the gigabytes or even terabytes, conducting an exhaustive search through the data is practically impossible. This necessitates the use of sophisticated computational strategies for the analysis and interpretation of sequencing data.
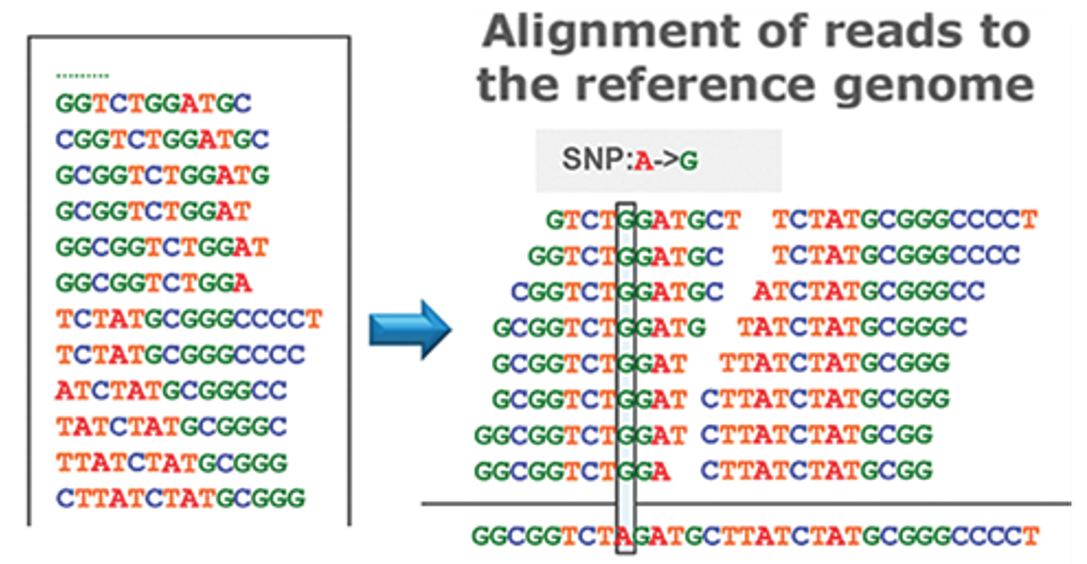 Figure 6: DNA analysis by short-read sequencing. Adapted from DOI:10.5772/66732
Figure 6: DNA analysis by short-read sequencing. Adapted from DOI:10.5772/66732
Given the vast amounts of data, searching for a single-letter mutation in a long sequence of letters is akin to looking for a needle in a haystack. Also, comparing the thousands of RNA molecules present within each cell across all different cells is a tremendous task. Dimensionality reduction is one approach for managing such large data volumes. This technique aims to reduce data size while preserving its essential properties—those that enable us to make relevant distinctions between different data samples. It involves representing phenomena captured in one dimension (e.g., a 3D representation) in a lower dimensional representation (e.g., a 2D representation). In medical contexts, these lower dimensional representations help to process and analyze information contained within the higher dimensional data. Dimensionality reduction is often used to create 2D visualizations of complex data, alongside clustering techniques that make found patterns visually relatable to us. Assuming that dimensionality reduction maintains the relative structure between data points, it allows us to group these points based on proximity into clusters, which likely represent cells of the same type (Fig. 7).
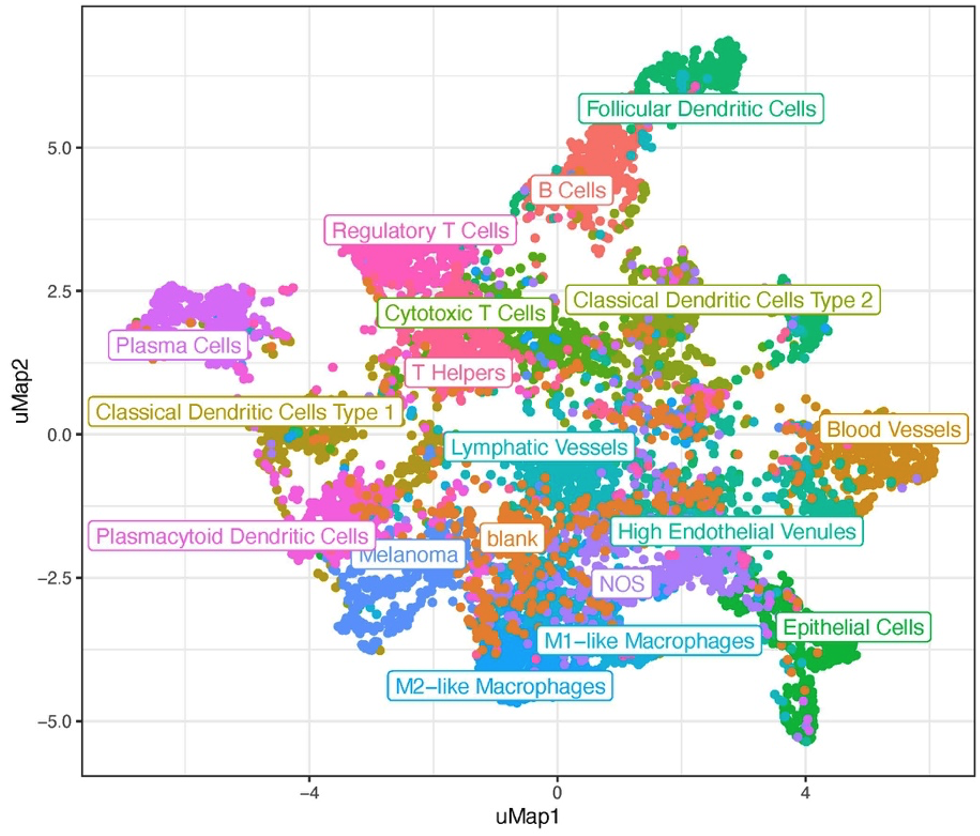 Figure 7: UMAP visualizing 18 different cell types identified after clustering. Adjusted from DOI:10.1158/0008-5472.CAN-22-0363
Figure 7: UMAP visualizing 18 different cell types identified after clustering. Adjusted from DOI:10.1158/0008-5472.CAN-22-0363
Exciting examples in this context come from one of our members, Prof. Stein Aerts and his team, who develop and apply machine learning models and tools to analyse multiple single-cell omics (aka multi-omics) datasets (see below for multi-omics). One of these tools is called SCENIC+, and aims to infer gene regulatory mechanisms underlying the identity of cell types. It does so by deploying a variety of machine learning methods that allow to reveal crucial differences in gene regulation between healthy cells and cancer cells or across different cancer cell states.
“Through the comparison of gene regulatory programs across cancers using SCENIC+ and deep learning models, we aim to find key regulators per cancer cell state and pave the way for discovering more specific biomarkers and new leads for therapeutic intervention.”
— Vasilis Konstantakos and Seppe De Winter
Spatial omics: because location matters
A tumor is composed of various cell types, including cancer cells, immune cells, cells from blood vessels, and connective tissue. Understanding the tumor’s biology requires knowledge not only of the intrinsic characteristics of each of these individual cells but also of their absolute and relative locations within the tissue. Spatial omics, akin to the methods previously described, analyze biological molecules but with a key distinction: they incorporate the spatial distribution of these molecules into their analysis. Therefore, the data collected per tissue spot can include sequencing data (strings of ATCG), chromatograms generated by mass spectrometry, or even dozens to hundreds of images from fluorescence microscopy (Fig. 8).
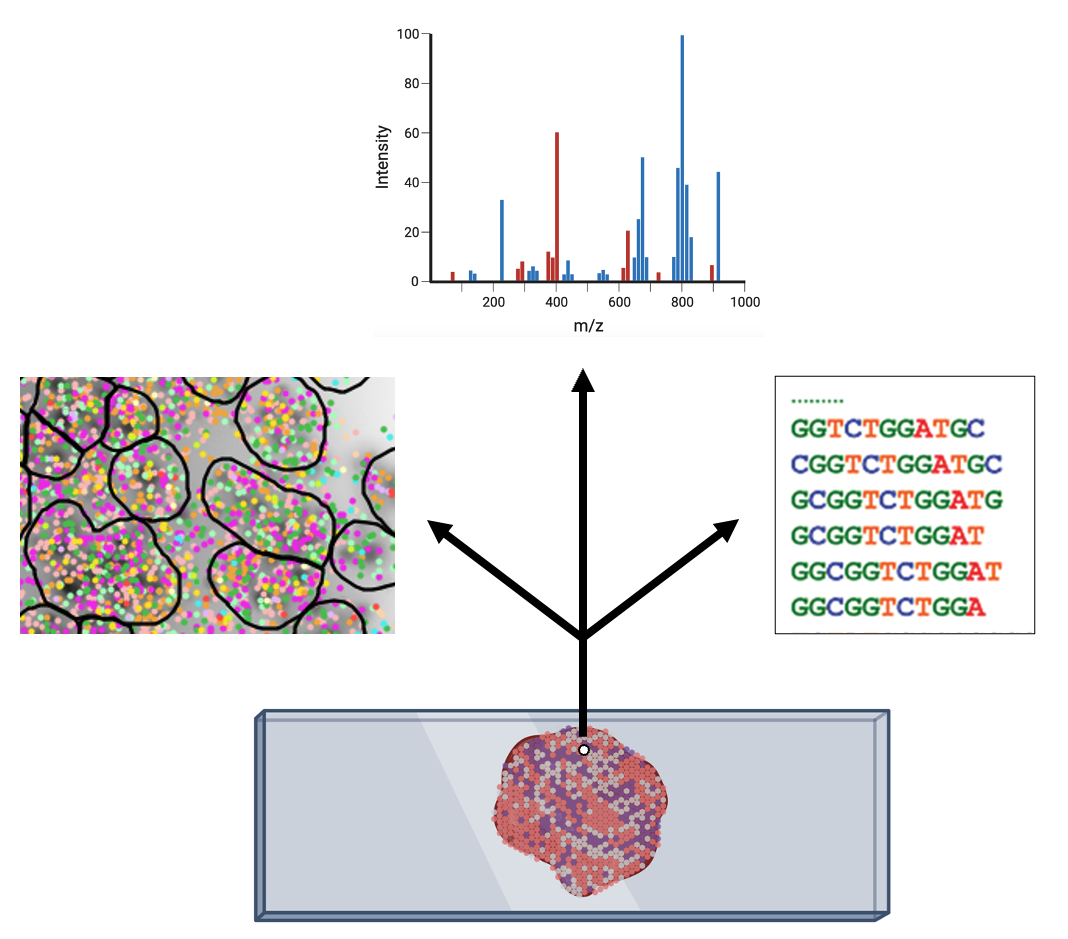 Figure 8: Depending on the technology, different types of data can be collected per spot within a tissue (Created with Biorender).
Figure 8: Depending on the technology, different types of data can be collected per spot within a tissue (Created with Biorender).
Detailed mapping of every cell within cancer tissue enables a deeper understanding of complex disease mechanisms and facilitates more accurate prognostic and predictive assessments regarding the effectiveness of cancer treatments. For instance, is it possible to associate the spatial distribution of tumor cells with their communication capabilities? Or can we correlate spatial arrangements of certain immune cell types with their ability to eradicate tumor cells? Creating a comprehensive spatial pathology map that encompasses all relevant cell types within the tumor is precisely the goal of researcher and pathologist Prof. Francesca Bosisio and her team (Fig. 9). In a study involving patients with metastatic melanoma, they identified spatial patterns that can predict how patients will respond to immunotherapy.
“The use of simple biomarkers is no longer suitable to predict the effect of novel therapeutic approaches – it’s the presence of multiple specific (immune) cell types, in the right location at the right time, that is becoming truly informative. This can only be inferred using far more complex technologies and data analysis tools than are currently available within the clinics”
— Prof. Francesca Bosisio
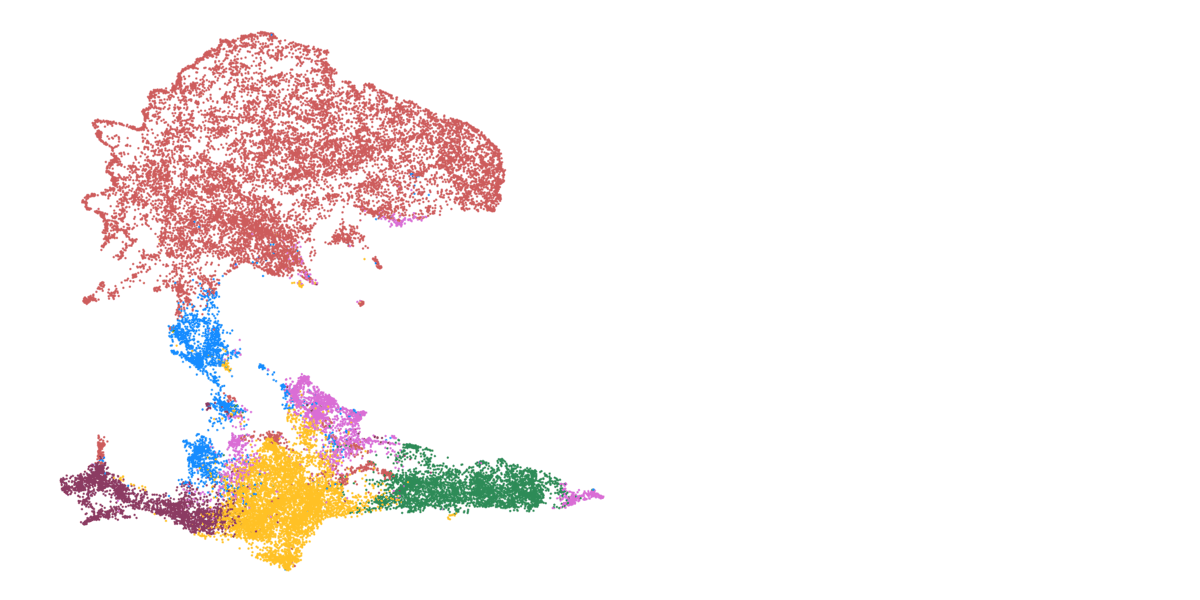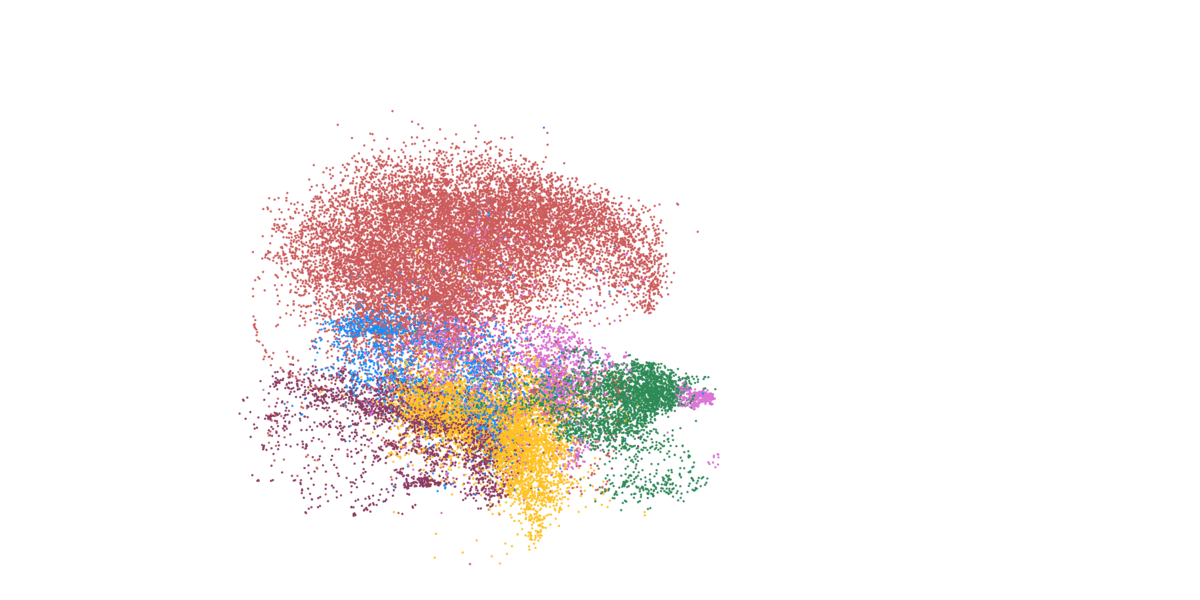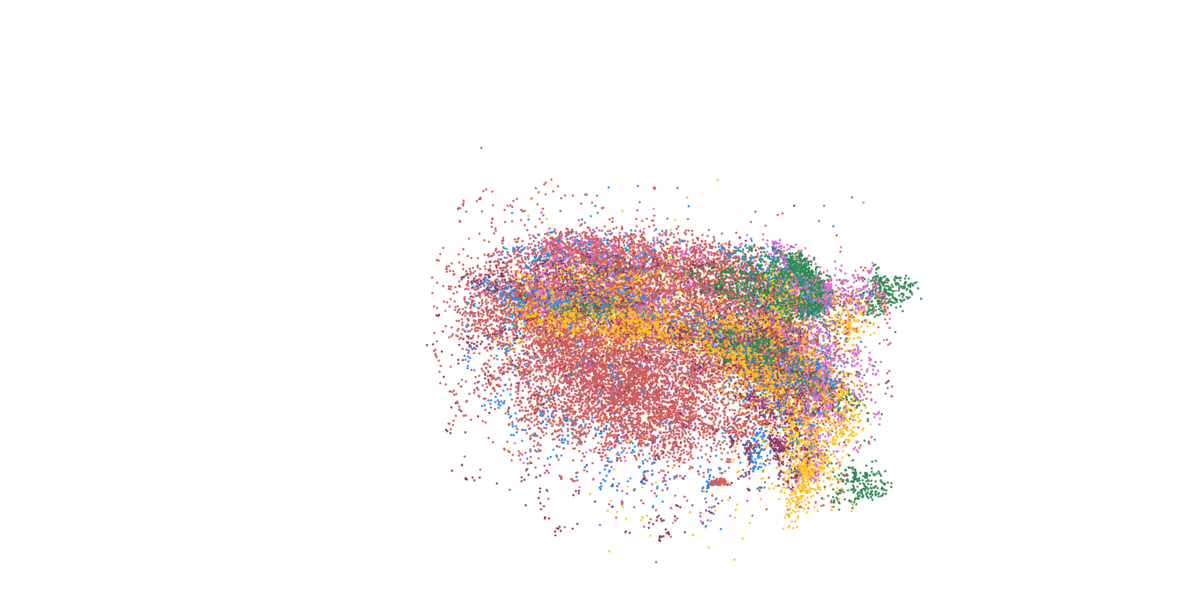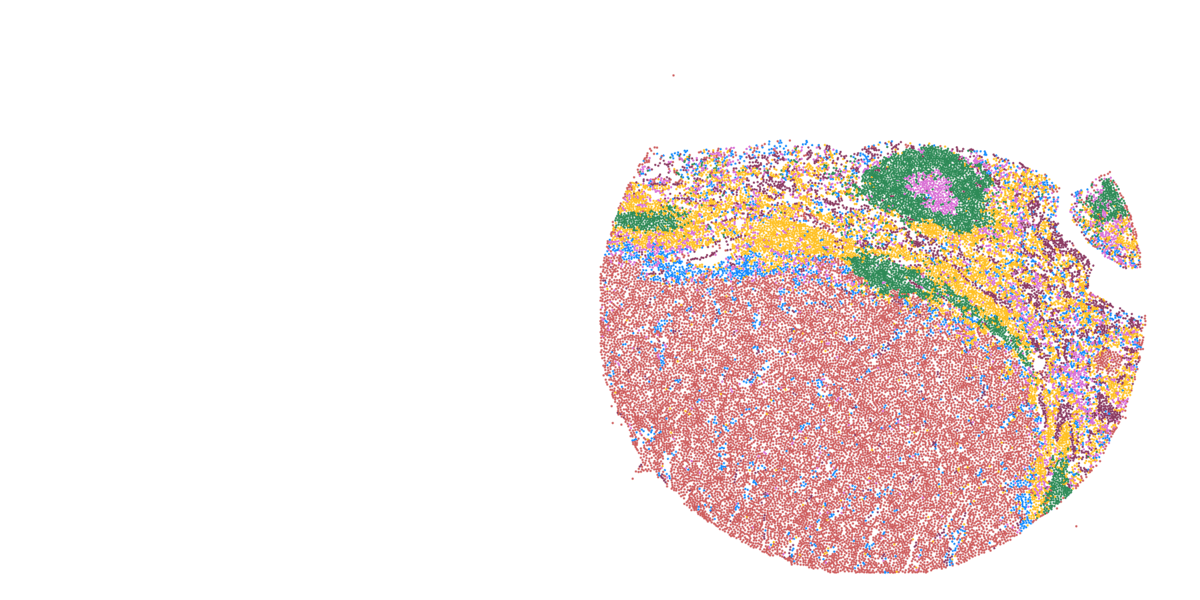 Figure 9: Cells, clustered in different cell types, mapped to their actual position within the tissue.
Figure 9: Cells, clustered in different cell types, mapped to their actual position within the tissue.
The challenges encountered in the analysis of single-cell data are also present in spatial omics studies, with the added complexity of incorporating spatial dimensions. Researchers explore the intricacies of spatial structures of cells and, for instance, specific patterns of gene expression within them, by employing medical imaging algorithms tailored to spatial datasets. As previously mentioned, these algorithms aim to detect patterns in the distributions of image data. However, the process is more complex. For example, spatial transcriptomics data may be represented by points in x,y or x,y,z coordinates. Converting these individual data points into image data can unnecessarily enlarge the data size and introduce unwanted artifacts. To circumvent these issues, researchers are exploring AI methods specifically designed to recognize structures and patterns in point cloud representations. Developing such tailored methods for each type of data modality holds the promise of a more precise understanding of tumor biology.
Single-cell and spatial multi-omics: a matter of integration
Given the multifaceted nature of cancer, analyzing just one omic layer may not capture the entire molecular dysfunction and thus the complexity of the disease. To address this, multi-omics technologies have been developed, enabling the integration of various omics data types into a joint analysis. This approach allows for the exploration of interactions and dependencies among different classes of biomolecules within individual cells and/or their spatial relationships. By combining data from single-cell and spatial analyses, researchers can gain insights into the multiple, often interrelated, molecular processes that malfunction within a tumor. This enhanced understanding of the cellular complexity and plasticity of tumors, coupled with the growing number of available therapies, underscores the necessity of identifying an increasing array of biomarkers and characteristics of the various cells involved. This is crucial for achieving accurate personalized diagnoses and, consequently, tailored treatments.
“Technologies to measure different properties of the vast spectrum of cells involved in cancer biology at the single-cell level within the tissue context are in continuous innovation but will only come into their own if they can be integrated. To achieve this, investment must not only be made in the necessary combinations of cutting-edge technology, there is also a great need for an accompanying digital platform capable of analyzing the large amounts of data, and to keep cancer diagnostics efficient, affordable, and sustainable in the long term.“
— Prof. Frederik De Smet
In single-cell and spatial omics, AI tools play a crucial role in designing experiments by identifying the most informative features for analysis, such as selecting specific genes, and aiding in data analysis through cell segmentation, clustering, and annotation. When it comes to multi-omics analyses, an additional set of AI-based methodologies becomes important for integrating varied datasets. For example, combining data from different imaging modalities and ACGT sequences is essential to understand the full biological picture. Machine learning models, such as autoencoders or neural networks, take as input various data types and learn to create joint embeddings. These are internal representations, neither images nor ACGT sequences to use the same examples, that connect different data modalities. For instance, an embedding might correlate a specific ACGT subsequence with a particular fluorescent signal in an image. After training these models, the next step involves analyzing their embeddings to find patterns, such as by transforming these embeddings into lower-dimensional representations for clustering and visual exploration. Such innovative data fusion strategies for the integration of multi-omic datasets is the focus of Prof. Alejandro Sifrim’s team. In a recent study involving further KU Leuven and UZ Leuven researchers, alongside other institutions and Aspect Analytics, a KU Leuven bioinformatics spin-off, the developed spatial multi-omics integration pipeline uncovered unique molecular patterns linked to malignant disease. Remarkably, it also identified precancerous regions that appeared morphologically normal (and thus unpredictable by the pathologist) but exhibited molecular abnormalities (Fig. 10).
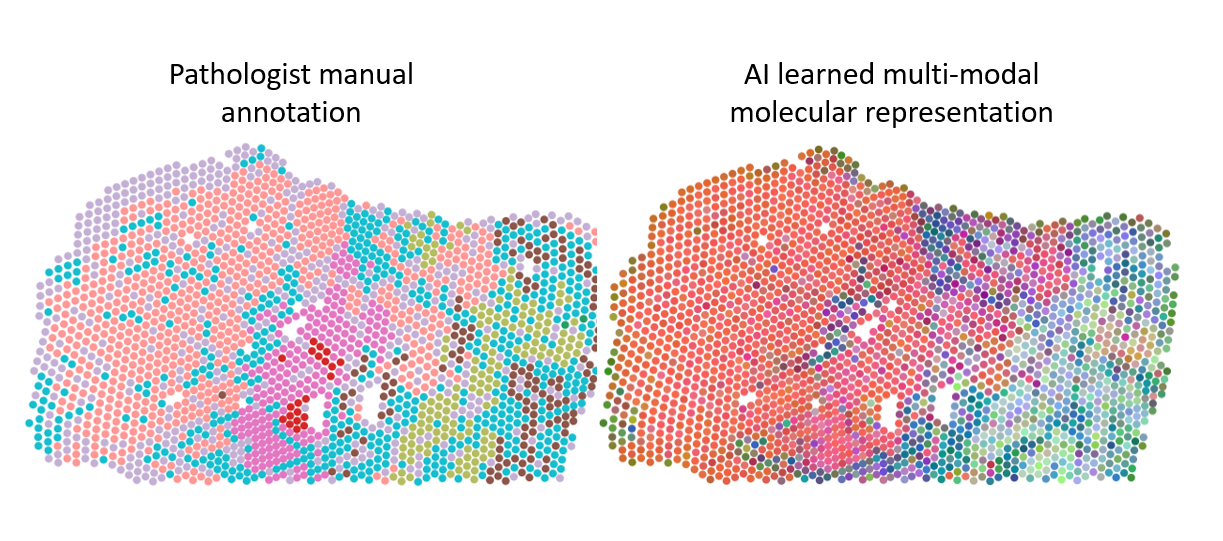 Figure 10: Different regions in prostate cancer tissue, manually annotated by a pathologist or clustered based on an integrated molecular representation generated with dedicated AI models. The multi-modal molecular representation is adding more granularity to the analyses, which allows better insight in tumor biology and highlight the potential to aid in histopathological annotation of ambiguous tissue areas.
Figure 10: Different regions in prostate cancer tissue, manually annotated by a pathologist or clustered based on an integrated molecular representation generated with dedicated AI models. The multi-modal molecular representation is adding more granularity to the analyses, which allows better insight in tumor biology and highlight the potential to aid in histopathological annotation of ambiguous tissue areas.
When multiple omics datasets acquired from the same sample are merged, the cell or a tissue feature can be leveraged as an anchor to ensure correct integration. The high costs of these multi-omics assays, however, limit the number of samples that can be analysed in one study. This often results in large amounts of omics data based on only a few patients, posing the risk of massive overfitting of data analyses to that small cohort. This can lead to results with limited validity considering the potentially more heterogenous nature of omics data when studying larger cohorts. And that’s exactly where orthogonal integration comes in – the integration of different, partially independent methods or experiments to further validate the obtained results. This challenging form of integration emerges as a trending topic since it brings together multiple omics datasets from different experiments and hence does not require anchored cells or matched features within the same sample.
“Very high-resolution technologies may cause substantial ‘loss of full transcriptome information’ and ‘loss of patient power’ due to costs of these technologies. Thus, orthogonal integration of multi-omics or spatial data across independent clinical cohorts is necessary to create biomarkers that work in clinical practice with cheaper technologies.”
— Prof. Abhishek D. Garg
The potential of this approach has already been demonstrated by multiple research lines led by KU Leuven researcher Prof. Abhishek Garg. Integration of different experimental datasets allowed him and his collaborators to define biomarkers to predict therapy response in several cancer types. In addition, they used multi-omics biomarkers to develop a so-called targeted assay to estimate patient survival and the risk of metastasis based on the serum of cancer patients. These kinds of targeted assays, which analyze pre-defined targets, instead of a full omics layer, offer a significant advantage in terms of clinical implementation compared to the previously described (multi-)omics analyses. Integrating data of different single-cell and spatial omics layers as well as different experiments will be crucial to develop a complete picture of what goes wrong within tumors on a molecular level. It will help us better understand how cancer develops from the early to very advanced stages, which molecular changes occur within the body following therapy and why some patients respond and others relapse. This, in turn, will be essential to advance the development of reliable biomarkers for diagnosis and therapy selection and the identification of innovative targets for therapies.
Concluding remarks
Personalized treatment and care are regarded as the holy grail of cancer care, necessitating a much deeper and more complex understanding of cancer biology. New technologies allow us to delve further into the human body, albeit at the expense of generating vast amounts of data. The pivotal role of artificial intelligence in this endeavor is unquestionable. However, the path to achieving this is not quite straightforward. It will demand considerable effort from a diverse set of stakeholders. This requires researchers and clinicians from various backgrounds to unite and dedicate their time to building the shared understandings and perspectives necessary for advancing complementary, yet distinct, expertise. Higher education institutions must provide the frameworks that facilitate such interdisciplinary collaboration on topics of significant societal importance – this is where institutes like LKI, LISCO, or Leuven.AI can play an essential role. By merging the invaluable data produced by the latest medical technologies with the vast potential of AI, we can enhance cancer care through earlier and more precise disease diagnosis, identification of new treatment targets, and better strategies for matching the right treatment to the right patient.
We’d like to thank the following colleagues for their input and feedback:
- Frederik De Smet (quote & input on financial sustainability and spatial multi-omics)
- Abhishek Garg (quote & input on orthogonal integration)
- Wouter Crijns (quote & input on medical imaging)
- Francesca Bosisio, Vasilis Konstantakos and Seppe De Winter (quote)
- Asier Antoranz (Figure 9)
- Alejandro Sifrim (Figure 10)
tags: deep dive, Focus on good health and well-being, Focus on UN SDGs















