
ΑΙhub.org
Unlocking the potential of entity-centric knowledge graphs: transforming healthcare and beyond

By Christos Theodoropoulos
Knowledge graphs (KGs) have become a cornerstone in organizing and utilizing information across various domains, from enhancing search engines to improving recommendation systems. KGs comprise nodes (entities) and edges (relations) that depict the knowledge within a specific field or a collection of domains. The potential of KGs to enable intricate reasoning and inference has been investigated across various endeavors, encompassing tasks such as information retrieval, and knowledge discovery. While KGs have come a long way, representing knowledge effectively remains a formidable challenge, especially in complex fields like healthcare and biomedicine. This article highlights our recent publication Representation Learning for Person or Entity-centric Knowledge Graphs: An Application in Healthcare (presented at K-CAP 2023) and explores the concept of entity-centric knowledge graphs, a relatively uncharted territory in the KG landscape, but one that holds immense promise in reshaping how we organize, access, and leverage data.
Understanding the challenge
Creating knowledge graphs involves mapping relationships between entities based on ontologies or schemas. However, when dealing with intricate domains like healthcare, this task becomes exceptionally challenging. The biomedical field is replete with complex, interdependent relationships between entities, a lack of standardization, and sparse data, making the traditional KG frameworks less effective. In theory, the traditional KGs can be queried to retrieve the information about a specific entity, however they are typically not centered around a specific entity or node of interest, such as an individual patient in healthcare scenarios, and are not particularly designed using an entity-centric knowledge schema. The maintenance and expansion of the KGs also impose challenges. This makes it difficult to extract the intricate updated details and relationships related to a single entity from the graph. Entity-centric KGs address this issue by focusing on central nodes and their associated information. Moreover, there is a critical demand for accurate depiction of information derived from various data sources related to individual patients. This necessity extends beyond its essential role in supporting routine hospital operations by healthcare providers; it is also indispensable in the realm of clinical research. This entails the development of innovative predictive models and the identification of novel crucial factors associated with unfavorable patient outcomes.
The potential of entity-centric KGs
Entity-centric KGs have the potential to revolutionize the representation of knowledge by zeroing in on a central entity, such as a patient, and capturing all facets of their data effectively. By doing so, they open the door to a wide range of downstream applications beyond conventional graph traversal and reasoning. These applications include generating graph embeddings and training graph neural networks (GNN) for predictive tasks. In our use-case, a comprehensive portrayal of individual patients should encompass not only their clinical attributes, such as diagnoses, medical procedures, and prescribed medications, but should also embrace a range of additional variables that hold potential as predictive indicators. These supplementary factors may encompass demographics, behavioral traits, and sociodemographic aspects, like smoking habits or employment status. The inclusion of this diverse array of information in models and analytical frameworks holds the promise of creating more refined tools for appraising treatment effectiveness and guiding these interventions toward the most pertinent patient cases.
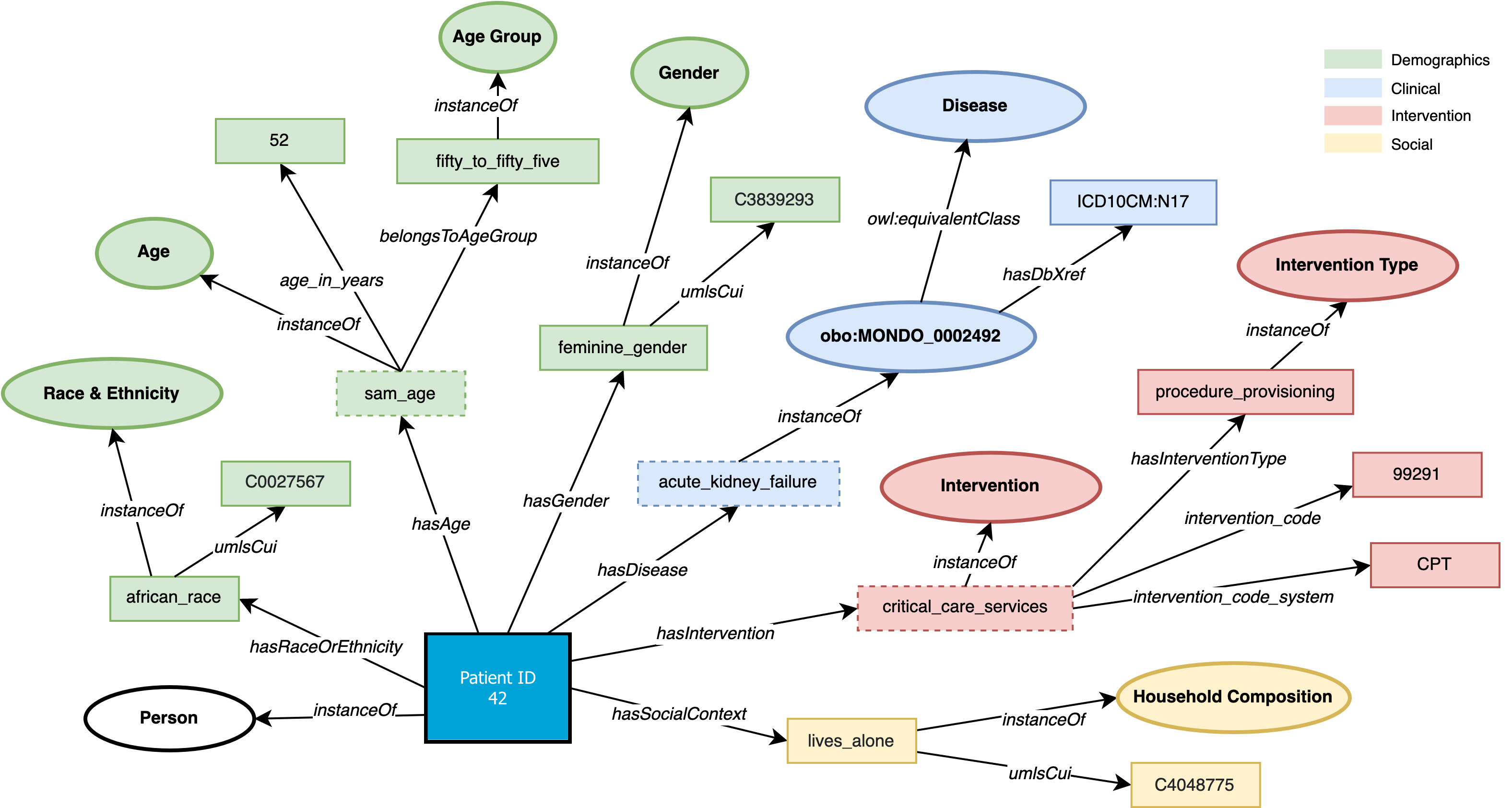 Figure 1: An illustration of a person-centric knowledge graph
Figure 1: An illustration of a person-centric knowledge graph
The innovative framework – use case
We introduce an end-to-end representation learning framework that extracts Personal Knowledge Graphs (PKGs) from structured and unstructured data, with a specific focus on healthcare. To guide the creation of these PKGs, we propose the star-shaped HSPO ontology that represents the various facets of a person’s health data. This ontology acts as a blueprint to construct a PKG that encapsulates all aspects related to a patient. The Health and Social Person-centric ontology (HSPO) has been created to provide a comprehensive perspective of an individual that spans various domains or aspects of their life. The HSPO establishes a knowledge schema for a star-shaped Personal Knowledge Graph, where a person serves as the central node, and associated characteristics or features (such as a computable phenotype) are connected to this central node. What makes this approach distinctive is its adaptability to continual expansion with additional domains and facets related to the individual. This expansion could include, among other things, clinical, demographic, and social factors in its initial version, and the potential for future versions to incorporate behavioral, biological, or genetic information. By representing a holistic view of individuals, encompassing new information generated not only within the traditional healthcare context but beyond, it is anticipated to unlock fresh insights that could revolutionize patient treatment approaches and service delivery methods.
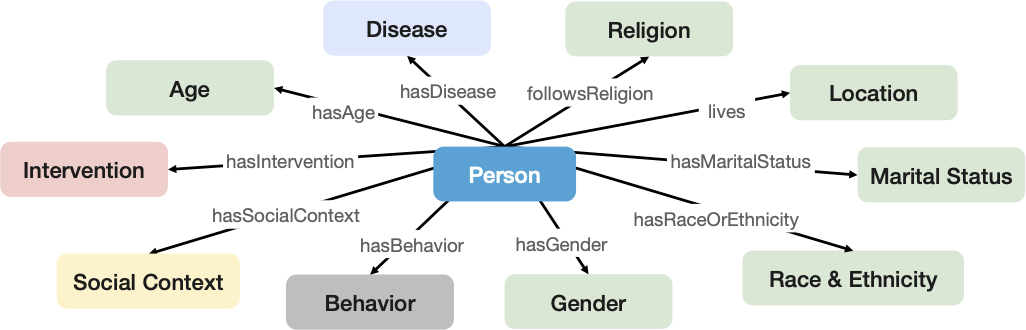 Figure 2: The main classes of the HSPO
Figure 2: The main classes of the HSPO
The heart of the framework lies in the utilization of graph neural networks, which enable the creation of compact representations. These representations are crucial for various predictive tasks, and our experiments encompass different levels of heterogeneity and explicitness within the data. We conducted experiments using two distinct GNN architectures, each with two variations (Figure 3). The variations stem from differences in the final layer of the model, which can be either convolutional or linear. The initial model, PKGSage, is founded on the Sage Graph Convolution Network, while the second model, PKGA, employs the Graph Attention Network (GAT) architecture. To evaluate the effectiveness of the proposed framework, we employed a readmission prediction task, which is a critical concern in healthcare. MIMIC-III stands out as an appropriate dataset for several reasons. It is well-suited not only due to its structured data and the diverse types of information it contains but also because of its patient population characteristics, particularly their reasons for hospital admission. It’s noteworthy that over 35% of the patients within this dataset were admitted for cardiovascular conditions, which have broad relevance in healthcare systems worldwide. Notably, a substantial percentage of heart failure patients face readmission within 30 days after their initial hospital discharge, with about half of them being readmitted within six months. These potentially preventable subsequent hospitalizations have been on the rise, prompting governments globally to target reducing 30-day readmissions as a long-standing goal to enhance healthcare quality (e.g., outcomes) and cut costs. In light of this, the experiments conducted and the knowledge graphs created in this study focus on patients admitted with cardiovascular conditions and aim at predicting potential 30-day readmissions. This predictive task can be extended to cover other medical conditions since readmissions serve as a widely accepted metric for evaluating success in healthcare. Moreover, the approach proposed in our paper, along with the associated open-source code repository, can be applied to other outcome metrics like specific events and mortality.
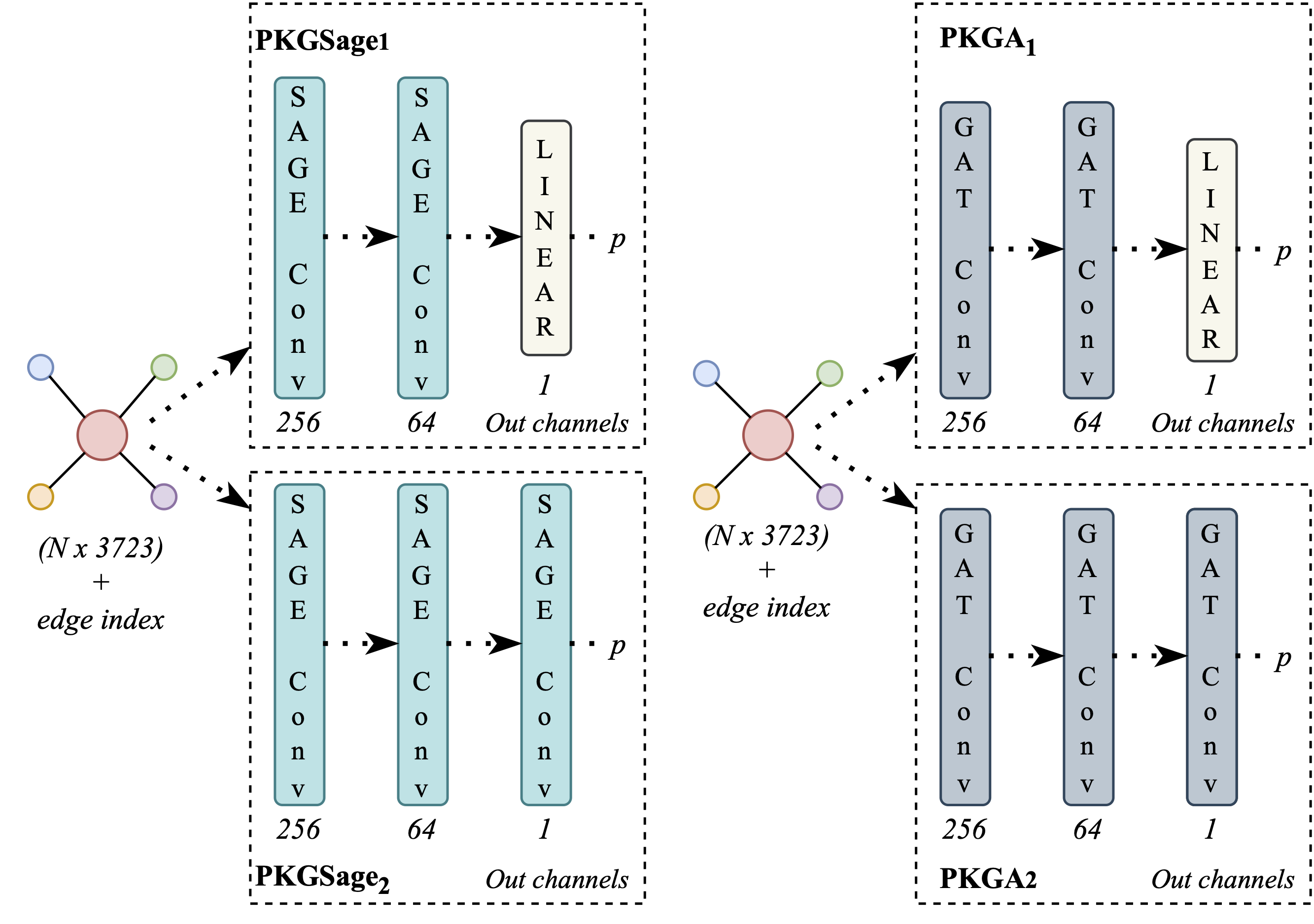 Figure 3: The model architectures of our study
Figure 3: The model architectures of our study
The results are promising, showing that the system we developed is stable, robust in the face of missing data, and outperforms several baseline machine learning classifiers. More precisely, an enhancement of 3.62% in F1-Score is observed. The best-performing model of the study achieves 62.16% accuracy and 68.06% F1-score. This success demonstrates the potential of entity-centric KGs in healthcare. The suggested end-to-end framework for person or entity-centric Knowledge Graphs (KGs) holds significance in both the technological landscape and various industries, as the framework is by design easily adaptable. While our primary focus is on the healthcare sector, it’s important to note that our approach is intentionally designed to be adaptable across different domains and isn’t confined to a specific final predictive task. The star-shaped KG approach and ontology we propose not only facilitate the meaningful integration of data from diverse sources and types, as already demonstrated, but also enable the exploration of new research questions. This sets the stage for the creation of a comprehensive and holistic 360-degree view of an individual, drawing from multiple data sources. Moreover, this approach can be extended to encompass patient cohorts or groups of individuals, providing valuable insights into a range of healthcare scenarios.
One of the key highlights of our framework is that it is open-sourced, allowing researchers, developers, and organizations to leverage this technology for their own applications and domains. We encourage collaboration and exploration of new use cases for entity-centric KGs. While our focus has been primarily on healthcare, the implications of entity-centric KGs extend far beyond this domain. They can be applied to various sectors, such as finance, e-commerce, and education, where understanding relationships and dependencies is essential.
Discussion and next steps
The approach offers scalability, allowing the creation of multiple personal knowledge graphs, one for each patient. This sets it apart from conventional methods that rely on very large general knowledge KGs, some of which have faced scalability challenges. The structure of traditional KGs, designed for querying and inferring knowledge, may not be well-suited for learning graph representations using graph neural networks due to their extensive size. Our experiments demonstrate that when heterogeneity is reduced through relation grouping, there is a positive impact on the results, provided that the PKG structure is generally suitable for learning patient representations with GNNs. We also note that the proposed framework exhibits a capacity for generalization even when the amount of data is limited.
Furthermore, the ontology design process can serve as a guide for developing other entity-centric ontologies, and the Health and Social Person-centric ontology (HSPO) is continuously expanding with new domains and facets. The open-sourced implementation for creating PKGs can be adapted and reused, with necessary adjustments, to extract entity-centric knowledge graphs in RDF or PyTorch Geometric applicable formats. Additionally, a wide range of predictive tasks using neural networks or knowledge graphs can be addressed, albeit constrained by data availability and annotation completeness.
We illustrate the applicability of this framework through a readmission prediction task using the PKGs and GNN architectures. Following this paradigm, other classification tasks, such as mortality prediction and clinical trial selection, can also be undertaken. The efficient representation of patients may be leveraged for clustering and similarity grouping applications.
In closing, we acknowledge that there are still challenges to address, including scalability, data integration, and privacy concerns. However, we firmly believe that the adoption of entity-centric KGs is the way forward in transforming knowledge representation and utilization. In conclusion, entity-centric knowledge graphs represent a ground-breaking approach to managing complex, interrelated data. In healthcare and beyond, this innovation promises to unlock new insights and applications, propelling knowledge representation to new heights. As the realm of entity-centric KGs continues to evolve, it is an exciting journey that holds great potential for discovery and progress.
Find out more
- Official Paper: Representation Learning for Person or Entity-centric Knowledge Graphs: An Application in Healthcare
- Pre-print: Representation Learning for Person or Entity-centric Knowledge Graphs: An Application in Healthcare
- Health & Social Person-centric Ontology (HSPO) – Official GitHub repository
- Leuven.AI Stories
This article is based on the work completed while I was a research intern at IBM Research Europe. The invaluable support and resources offered by the organization significantly contributed to the development and completion of the research. Special thanks are extended to my esteemed colleagues, Joao Bettencourt-Silva and Natalia Mulligan, whose collaboration and expertise enriched the content and analysis presented in this work. Their dedication and insightful contributions played a crucial role in shaping the outcomes of the research project. I am also grateful to Thaddeus Stappenbeck from Cleveland Clinic for sharing his insights and expertise. A special thanks is reserved for my supervisor, Marie-Francine Moens, whose continuous support and encouragement were pivotal in motivating me to pursue this internship and navigate the challenges that came with it. Her guidance and mentorship have been instrumental in my professional development.
tags: deep dive











