
ΑΙhub.org
Generating a biomedical knowledge graph question answering dataset

By Xi Yan
The biomedical domain is a complex network of interconnected knowledge, encompassing genetics, diseases, drugs, and biological processes. While knowledge graphs (KGs) excel at organizing and linking this information, their complexity often makes them difficult for users to query. Ideally, users should be able to ask questions in natural language and receive precise answers directly from the KG, without needing specialized query expertise. However, enabling deep learning-based systems to query KGs using natural language remains a major challenge. Existing biomedical knowledge graph question answering (BioKGQA) datasets are small and limited in scope, typically containing only a few hundred question answering (QA) pairs. This scarcity of data hinders the development of robust and scalable QA systems, which are essential for critical applications such as clinical decision support, personalized medicine, and drug discovery.
PrimeKGQA addresses these challenges with a novel, scalable approach to dataset generation, harnessing the power of large language models (LLMs). Built on PrimeKG—a precision medicine-oriented knowledge graph that integrates data from 20 of the most-cited biomedical databases spanning ten biological scales, including genes, diseases, and drugs—PrimeKGQA leverages a generalizable, scalable, and training-free data generation framework. Using few-shot learning with LLMs, the framework transforms KG subgraphs (based on network motifs, see Figure 1) into SPARQL queries, which are subsequently converted into natural language question-answer pairs. The resulting PrimeKGQA dataset encompasses a wide array of biomedical concepts and reasoning complexities, ranging from straightforward factual queries to intricate multi-hop reasoning paths, providing a comprehensive resource for advancing biomedical question-answering systems.
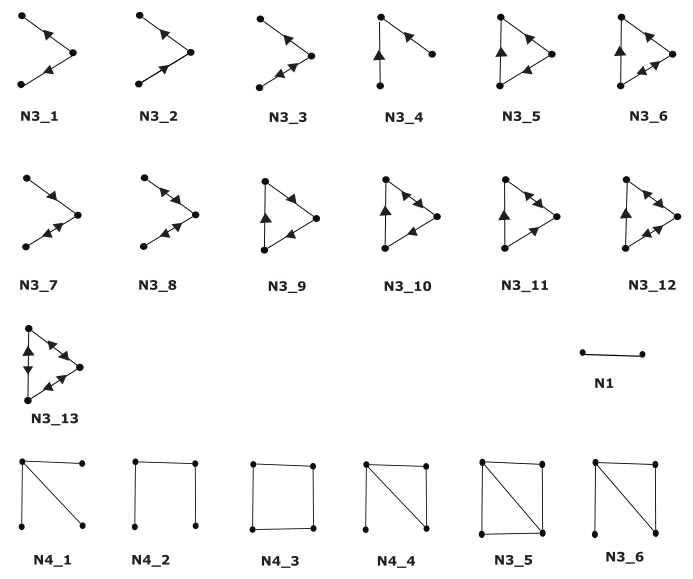 Figure 1: All types of network motifs for graphs with node numbers from two to four. N3_1 stands for “node number 3 subgraph type 1”. Note that for 3-node-subgraphs, we discard N3_5, N3_6, N3_9, N3_10, N3_11, N3_12 and N3_13.
Figure 1: All types of network motifs for graphs with node numbers from two to four. N3_1 stands for “node number 3 subgraph type 1”. Note that for 3-node-subgraphs, we discard N3_5, N3_6, N3_9, N3_10, N3_11, N3_12 and N3_13.
PrimeKGQA stands out not just for its size but also for its comprehensiveness. With 83,999 QA pairs, it is 1,000 times larger than the next largest BioKGQA dataset. The dataset includes questions generated from 2- to 4-node subgraphs, offering a balanced mix of simple and complex reasoning tasks. The questions are evaluated for linguistic correctness, semantic fidelity, and grammatical accuracy, ensuring a strong alignment with the biomedical KG facts it represents.
The creation of PrimeKGQA follows an innovative pipeline. 1. Subgraph Sampling: Subgraphs from PrimeKG are extracted based on network motifs, ranging from simple 2-node structures to more complex 4-node configurations. 2. SPARQL Validation: SPARQL queries are used to validate the answers extracted from the subgraphs, ensuring correctness of the answer. 3. Question Generation: Pre-trained language models like GPT3, Mistral, and LLaMA are prompted to generate natural language questions based on the subgraph structures and validated answers. A detailed pipeline could be found in figure 2.
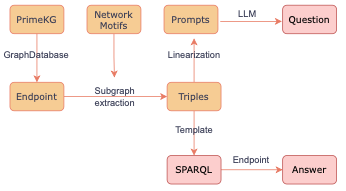 Figure 2. Our pipeline for automatic generation of PrimeKGQA. The Pink blocks are the composing elements of the dataset, i.e., natural question, SPARQL, Correct Answer from the KG.
Figure 2. Our pipeline for automatic generation of PrimeKGQA. The Pink blocks are the composing elements of the dataset, i.e., natural question, SPARQL, Correct Answer from the KG.
LLM-generated data is known to suffer from hallucination, which in our task is evaluated across three dimensions: grammaticality, consistency (whether the question and answer correspond), and coverage (whether the question and SPARQL query align). To assess this, we use both automatic and manual evaluation methods. Established benchmarks like BLEU, ROUGE, and METEOR are employed to measure linguistic quality, while LLM-based metrics like BERTScore ensure semantic alignment. Additionally, domain experts evaluate the samples for grammaticality, consistency, and coverage, providing human validation of the dataset’s overall quality.
While PrimeKGQA establishes a new benchmark for BioKGQA datasets, the work is far from complete. There are still opportunities for improvement, such as post-editing and correcting problematic questions. Additionally, no models have yet been tested on this dataset. Future work will focus on: refining the question generation process to capture more nuanced and exploratory queries, and using PrimeKGQA to benchmark existing QA systems to evaluate their effectiveness in real-world biomedical tasks.
Want to explore PrimeKGQA for yourself? The dataset and models are available on GitHub.
Read the work in full
Bridging the Gap: Generating a Comprehensive Biomedical Knowledge Graph Question Answering Dataset, Xi Yan, Patrick Westphal, Jan Seliger, and Ricardo Usbeck, ECAI 2024.
This research was presented at ECAI 2024.
tags: deep dive, ECAI, ECAI2024










